import os
import sys
base_dir = '/home/abzoghbi/data/swift_j2127.4p5654/nustar_re_analysis'
if not base_dir in sys.path: sys.path.insert(0, base_dir)
from helpers import *
%load_ext autoreload
%autoreload 2
The autoreload extension is already loaded. To reload it, use:
%reload_ext autoreload
os.chdir(base_dir)
wdir = 'data/timing'
os.system('mkdir -p %s'%wdir)
os.chdir(wdir)
nu_obsids = np.array(['60001110002', '60001110003', '60001110005', '60001110007', '60402008002',
'60402008004', '60402008006', '60402008008', '60402008010'])
3-79 keV Total light curve
#!rm lc_1b_512.npz
loc_info, nen, dt = [base_dir, '1b'], 1, 512
LC = read_lc(loc_info, nu_obsids, dt, nen, combine_ab=True)
#!rm lc_22l3_512_bgd.npz
loc_info, nen, dt = [base_dir, '22l3'], 22, 512
LCb = read_lc(loc_info, nu_obsids, dt, nen, combine_ab=True, bgd=True)
LC = remove_high_bgd(LC, LCb)
reading data from lc_1b_512.npz ..
reading data from lc_22l3_512_bgd.npz ..
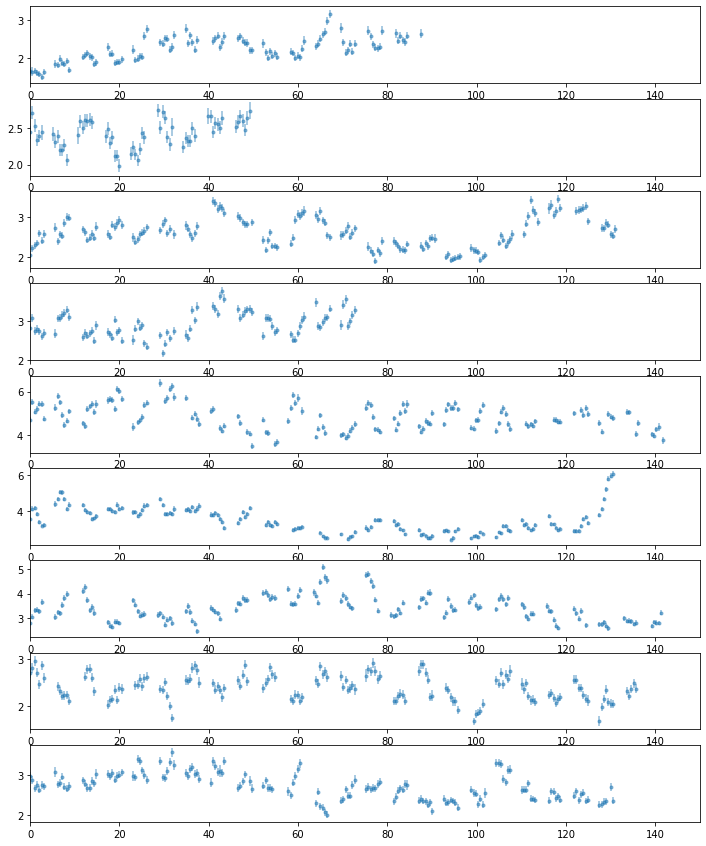
# plot light curves
nlc = len(LC[0])
fig, ax = plt.subplots(nlc, 1, figsize=(12, 15))
for ilc,lc in enumerate(LC[0]):
ax[ilc].errorbar((lc[0] - lc[0][0])/1e3, lc[1], lc[2], fmt='o', ms=3, alpha=0.5)
ax[ilc].set_xlim([0, 150])
dates = []
for o in nu_obsids:
with pyfits.open(f'{base_dir}/data/nustar/{o}_p/lc/1b/lc_512__a__1.lc') as fp:
dates.append(fp[1].header['date-obs'])
dates = TT.Time(dates)
# write lc
os.system('mkdir -p psd')
txt = ''
t0 = 0
for ilc,lc in enumerate(LC[0]):
tt = lc[0] - lc[0][0] + t0
t0 = tt[-1] + 1e4
tmjd = (dates[ilc] + TT.TimeDelta(lc[0] - lc[0][0], format='sec')).mjd
txt += '\ndescriptor t_%d rate_%d,+- tt_%d tmjd_%d\n'%(ilc+1,ilc+1,ilc+1, ilc+1)
txt += '\n'.join(['%10.10g %10.3g %10.3g %10.10g %10.7g'%(t,r,re,t_,_tm) for t,r,re,t_,_tm in
zip(lc[0]-lc[0][0], lc[1], lc[2], tt, tmjd)])
with open('psd/lc_%g.plot'%(dt), 'w') as fp: fp.write(txt)
PSD
# get segments #
tlen = 70
Lc, LcIdx = split_LC_to_segments(LC, tlen*1e3, plot=False)
# frequency bins
fqL, fqd = get_fq_bins(Lc[0], dt, mode=1, Nfq=10, nyquist=0.5)
psd_all_f1 = calculate_psd(Lc[0], fqL, 'psd/psd_all_f1.npz', errors=True)
nfq: 12
fqL: 5.71089e-06 8.27373e-06 2.39733e-05 3.47317e-05 5.03179e-05 7.28987e-05 0.000105613 0.000153008 0.000221672 0.00032115 0.000465271 0.000674067 0.00195312
126.083 | 4.92 6.15 3.9 4.6 4.65 3.68 2.98 3 1.86 0.79 0.069 -3.85 | 5.84e-07
** done **
126.083 | 4.92 6.15 3.9 4.6 4.65 3.68 2.98 3 1.86 0.79 0.069 -3.85 | 2.6e-07
** done **
## errors for param 0 ##
126.083 125.582 4.92058 6.58456 1.00218
## errors for param 1 ##
126.083 125.581 6.15142 6.43212 1.00478
## errors for param 2 ##
126.083 125.579 3.90129 4.69842 1.00795
## errors for param 3 ##
126.083 125.585 4.5973 4.95267 0.994948
## errors for param 4 ##
126.083 125.583 4.64735 4.90512 0.998878
## errors for param 5 ##
126.083 125.584 3.68308 3.93645 0.997221
## errors for param 6 ##
126.083 125.588 2.98128 3.23562 0.99037
## errors for param 7 ##
126.083 125.585 3.00326 3.18251 0.996679
## errors for param 8 ##
126.083 125.584 1.8554 2.083 0.998198
## errors for param 9 ##
126.083 125.578 0.790132 1.08586 1.00964
## errors for param 10 ##
126.083 125.583 0.0689913 0.337982 0.999747
## errors for param 11 ##
126.083 125.587 -3.84914 -2.51337 0.991134
********************
4.92 6.15 3.9 4.6 4.65 3.68 2.98 3 1.86 0.79 0.069 -3.85
1.66 0.281 0.797 0.355 0.258 0.253 0.254 0.179 0.228 0.296 0.269 1.34
********************
# Another frequency binning
fqL, fqd = get_fq_bins(Lc[0], dt, mode=1, Nfq=14, nyquist=0.5)
psd_all_f2 = calculate_psd(Lc[0], fqL, 'psd/psd_all_f2.npz', errors=True)
nfq: 16
fqL: 5.71089e-06 7.5414e-06 1.99173e-05 2.63013e-05 3.47317e-05 4.58641e-05 6.05649e-05 7.99777e-05 0.000105613 0.000139465 0.000184167 0.000243198 0.00032115 0.000424088 0.000560021 0.000739524 0.00195312
128.026 | -30 6.34 5.07 3.89 4.84 4.45 4.34 3.67 2.78 3.22 2.69 1.82 0.568 0.512 -0.388 -18 | 7.99e-17
** done **
128.026 | -30 6.34 5.07 3.89 4.84 4.45 4.34 3.67 2.78 3.22 2.69 1.82 0.568 0.512 -0.388 -18 | 7.99e-17
** done **
## errors for param 0 ##
## errors for param 1 ##
128.026 127.526 6.33941 6.60503 1.00028
## errors for param 2 ##
128.026 127.526 5.07 5.76921 1.00044
## errors for param 3 ##
128.026 127.523 3.88629 4.87066 1.00664
## errors for param 4 ##
128.026 127.528 4.83719 5.22782 0.997309
## errors for param 5 ##
128.026 127.527 4.45166 4.81104 0.998306
## errors for param 6 ##
128.026 127.524 4.34309 4.64777 1.00486
## errors for param 7 ##
128.026 127.523 3.67401 3.95136 1.00647
## errors for param 8 ##
128.026 127.53 2.78425 3.10457 0.992306
## errors for param 9 ##
128.026 127.525 3.21512 3.43387 1.00314
## errors for param 10 ##
128.026 127.524 2.69011 2.89714 1.00421
## errors for param 11 ##
128.026 127.53 1.81543 2.05371 0.991899
## errors for param 12 ##
128.026 127.521 0.567792 0.970136 1.00973
## errors for param 13 ##
128.026 127.527 0.51227 0.783755 0.998757
## errors for param 14 ##
128.026 127.525 -0.387784 -0.0284093 1.00141
## errors for param 15 ##
********************
-33.9 6.34 5.07 3.89 4.84 4.45 4.34 3.67 2.78 3.22 2.69 1.82 0.568 0.512 -0.388 -18
9.5 0.266 0.699 0.984 0.391 0.359 0.305 0.277 0.32 0.219 0.207 0.238 0.402 0.271 0.359 9.5
********************
# Analytical fit with PL and BPL
psd_all_f = calculate_psd(Lc[0], fqL, 'psd/psd_all_f.npz', do_bin=0, do_pl=1, do_bpl=1, errors=True)
80.5587 | -13.3 -1.8 | 7.1e-07
** done **
80.5587 | -13.3 -1.8 | 2.82e-11
** done **
## errors for param 0 ##
80.5587 80.0558 -13.3166 -12.8427 1.00591
## errors for param 1 ##
80.5587 80.0634 -1.79922 -1.74445 0.990602
********************
-13.3 -1.8
0.474 0.0548
********************
120.181 | -5.47 -3.86 -8.28 | 1.79e-05
** done **
120.181 | -5.47 -3.86 -8.28 | 1.4e-05
** done **
## errors for param 0 ##
120.181 119.685 -5.46904 -5.35771 0.990184
## errors for param 1 ##
120.181 119.677 -3.86465 -3.46622 1.00745
## errors for param 2 ##
120.181 119.681 -8.27844 -8.15637 0.999052
********************
-5.47 -3.86 -8.28
0.111 0.398 0.122
********************
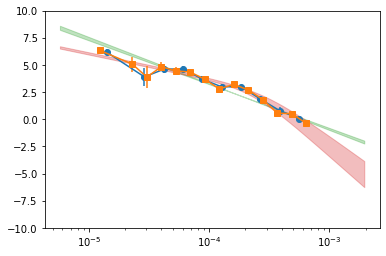
# plot the psd #
pBin = psd_all_f1['p_Bin']
plt.errorbar(pBin[0][1:-1], pBin[1][1:-1], pBin[2][1:-1], fmt='o-')
pBin = psd_all_f2['p_Bin']
plt.errorbar(pBin[0][1:-1], pBin[1][1:-1], pBin[2][1:-1], fmt='s-')
mPL = psd_all_f['m_pl']
plt.fill_between(mPL[0], mPL[1]-mPL[2], mPL[1]+mPL[2], alpha=0.3, color='C2')
mBPL = psd_all_f['m_bpl']
plt.fill_between(mBPL[0], mBPL[1]-mBPL[2], mBPL[1]+mBPL[2], alpha=0.3, color='C3')
plt.xscale('log')
plt.ylim([-10, 10])
(-10, 10)
# write psd_all #
txt = 'descriptor fq_f1 psd_f1,+-\n'
txt += '\n'.join(['%g %g %g'%(z[0], z[1], z[2]) for z in psd_all_f1['p_Bin'][:,1:-1].T])
txt += '\ndescriptor fq_f2 psd_f2,+-\n'
txt += '\n'.join(['%g %g %g'%(z[0], z[1], z[2]) for z in psd_all_f1['p_Bin'][:,1:-1].T])
txt += '\ndescriptor fq_pl psd_pl,+-\n'
txt += '\n'.join(['%g %g %g'%(z[0], z[1], z[2]) for z in psd_all_f['m_pl'].T])
txt += '\ndescriptor fq_bpl psd_bpl,+-\n'
txt += '\n'.join(['%g %g %g'%(z[0], z[1], z[2]) for z in psd_all_f['m_bpl'].T])
with open('psd/psd_all.plot', 'w') as fp: fp.write(txt)
p,pe = psd_all_f1['p_Bin'][1:,1:-1]
fqL = psd_all_f1['fqL'][1:-1]
az.misc.write_pha_spec(fqL[:-1], fqL[1:], p, pe, 'psd_f1')
p,pe = psd_all_f2['p_Bin'][1:,1:-1]
fqL = psd_all_f2['fqL'][1:-1]
az.misc.write_pha_spec(fqL[:-1], fqL[1:], p, pe, 'psd_f2')
os.system('mkdir -p psd/pha')
os.system('mv *pha *rsp *xsp psd/pha')
psd_f1.pha was created successfully
psd_f2.pha was created successfully
0