import os
import sys
base_dir = '/home/abzoghbi/data/swift_j2127.4p5654/nustar_re_analysis'
if not base_dir in sys.path: sys.path.insert(0, base_dir)
from helpers import *
%load_ext autoreload
%autoreload 2
os.chdir(base_dir)
wdir = 'data/timing'
os.system('mkdir -p %s/lag'%wdir)
os.chdir(wdir)
nu_obsids = np.array(['60001110002', '60001110003', '60001110005', '60001110007', '60402008002',
'60402008004', '60402008006', '60402008008', '60402008010'])
#!rm lc_22l3_512.npz
loc_info, nen, dt = [base_dir, '22l3'], 22, 512
LC = read_lc(loc_info, nu_obsids, dt, nen, combine_ab=True)
#!rm lc_22l3_512_bgd.npz
loc_info, nen, dt = [base_dir, '22l3'], 22, 512
LCb = read_lc(loc_info, nu_obsids, dt, nen, combine_ab=True, bgd=True)
LC = remove_high_bgd(LC, LCb)
reading data from lc_22l3_512.npz ..
reading data from lc_22l3_512_bgd.npz ..
# average counts per bin #
txt = '\n'.join([' '.join(['%8.4g'%(l[1].mean()*dt) for l in lc]) for lc in LC])
print(txt)
33.5 40.17 41.4 48.36 91.71 64.85 58.67 36.4 44.19
41.37 49.85 54.18 63.63 116.6 79.83 73.23 47.57 56.68
54.66 66.92 72.2 83.11 156.6 107.1 101.9 62.04 77.98
57.78 72.51 74.98 87.9 155.1 107.1 102.1 64.67 78.53
49.26 58.26 64.74 73.18 132.9 94.03 86.33 55.96 68.06
58.08 71.51 76.28 89.05 156.4 107.1 102.2 66.98 81.55
61.2 72.68 80.47 89.09 156.8 111.3 105 71.16 84.51
74.74 86.35 97.36 105.2 182.7 131.2 123.6 84.09 101.3
69.28 79.38 86.56 97.72 165.5 120.4 112.8 76.94 90.29
63.14 76.12 81.98 91.67 152.2 112.1 104.7 72.22 84.63
51.71 60.75 66.5 75.92 121.8 86.5 84.35 57.03 69.46
53.33 63.99 68.82 75 121.1 86.63 84.22 59.21 70.71
53.35 60.36 66.65 74.99 115.3 83.44 81.05 58.84 68.36
69.63 79.44 85.82 95.91 143.6 105.4 104 76.96 87.22
52.8 65.81 70.08 75.11 111.5 79.41 80.38 61.22 69.92
44.34 51.31 56.89 61.18 85.74 62.54 62.75 49 58.29
34.55 42.1 44.58 46.11 63.67 48.62 47.44 38.31 43.68
25.54 29.19 32.42 32.8 41.72 33.67 35.28 27.73 31.83
35.22 39.32 42.8 44.01 50.85 42.72 43.74 36.99 40.73
15.51 15.12 15.75 14.79 17 14.21 15.88 14.58 14.58
11.93 11.19 10.11 8.89 11.21 9.62 10.91 9.911 8.34
15.18 9.826 9.52 7.557 9.457 10.23 12.89 10.81 8.771
# plot light curves
ie = 21
nlc = len(LC[ie])
fig, ax = plt.subplots(nlc, 1, figsize=(12, 10))
for ilc,lc in enumerate(LC[ie]):
ax[ilc].errorbar((lc[0] - lc[0][0])/1e3, lc[1], lc[2], fmt='o', ms=3, alpha=0.5)
ax[ilc].set_xlim([0, 150])
plt.tight_layout()

ebins, dt = ('3 3.3 3.6 4 4.4 4.8 5.2 5.7 6.3 6.9 7.6 8.3 9.1 10 '
'11.7 13.8 16.2 19 22 31 42 58 79'), 512
tlen = 70
Lc, LcIdx = split_LC_to_segments(LC, tlen*1e3, plot=False)
_k: Reproduce lags in Fig 3 and 4 in Kara+14: <4e-5 4.5e-4
fqL = np.array([8e-6, 1e-5, 4e-5, 4.5e-4, 2e-3])
iEn = [[0,1], [2,3], [4,5], [6,7], [8,9], [10,11], [12,13], [14,15], [16,17], [18,19], [20,21]]
lag_k = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3b_k.npz', iEn=iEn, mcmc=[-4, 2000], logmod=False)
cache file lag/lag_22l3b_k.npz found. Reading ...!
lag_ka = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3b_ka.npz', iEn=iEn, iLc=[0,1,2,3],
mcmc=[-4, 2000], logmod=False)
cache file lag/lag_22l3b_ka.npz found. Reading ...!
lag_kb = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3b_kb.npz', iEn=iEn, iLc=[4,5,6,7,8],
mcmc=[-4, 2000], logmod=False)
cache file lag/lag_22l3b_kb.npz found. Reading ...!
# process mcmc from fqlag and plot histograms
lagMC_k = proc_lag_mcmc('lag_22l3b_k')
lagMC_ka = proc_lag_mcmc('lag_22l3b_ka')
lagMC_kb = proc_lag_mcmc('lag_22l3b_kb')
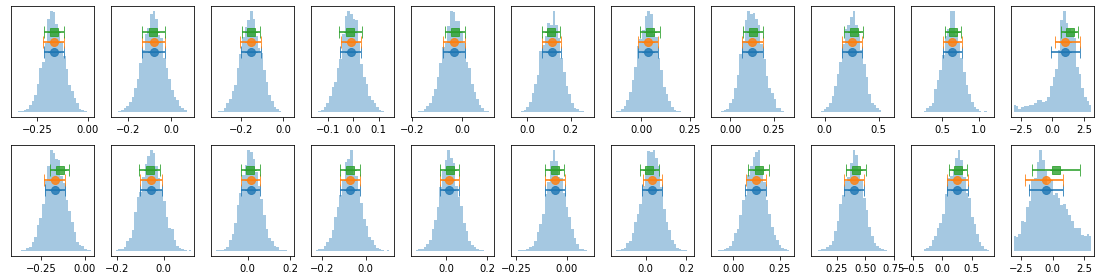
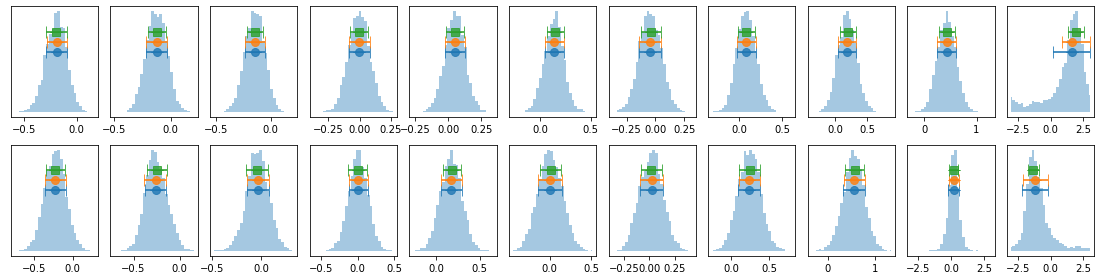
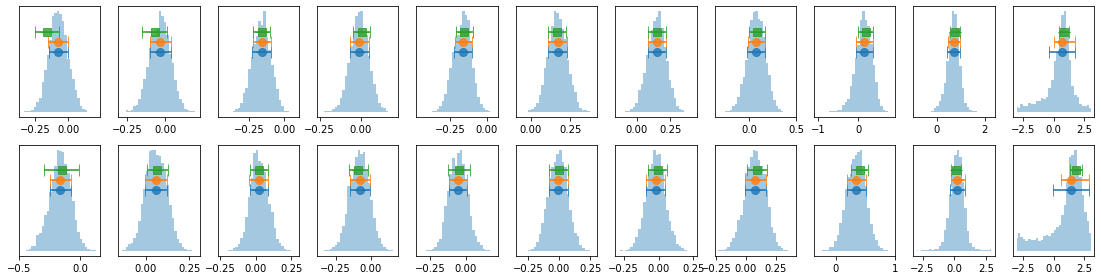
# plot lag-energy data from the mcmc chains #
for x in ['a', 'b', '']:
exec('plot_lag(lagMC_k%s)'%x)
exec('write_lag(lagMC_k%s, "_22l3bMC_k%s", pha=True)'%(x,x))
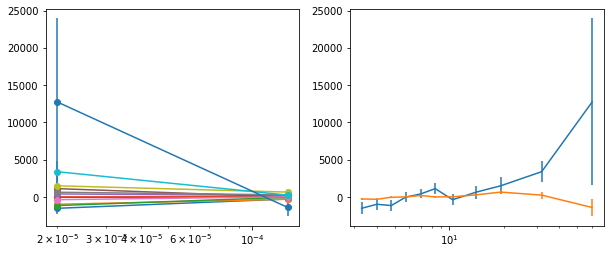
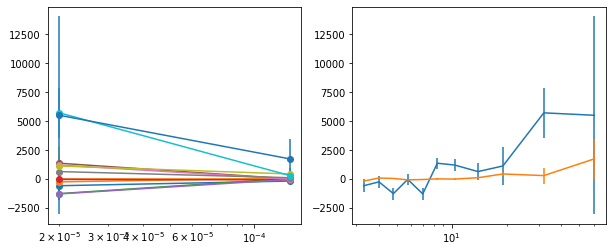
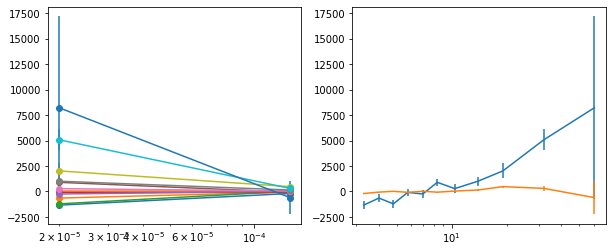
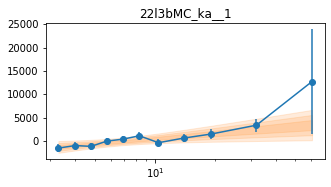
# Model the PHA data from fqlag mcmc with xspec
os.chdir('%s/%s/lag/pha'%(base_dir, wdir))
for x in ['a', 'b', '']:
fit_pha_with_loglin('22l3bMC_k%s__1'%x, recalc=1)
fit_pha_with_loglin('22l3bMC_k%s__2'%x, recalc=1)
os.chdir('%s/%s'%(base_dir, wdir))
chains for 22l3bMC_ka__1
chains for 22l3bMC_ka__2
chains for 22l3bMC_kb__1
chains for 22l3bMC_kb__2
chains for 22l3bMC_k__1
chains for 22l3bMC_k__2
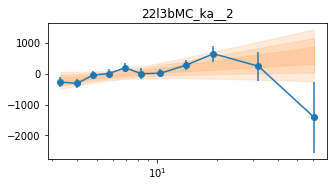
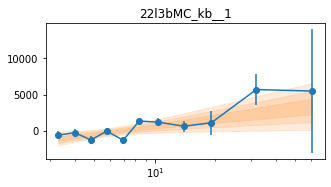
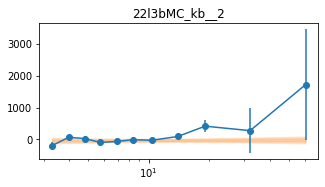
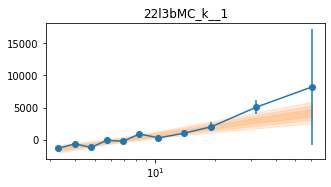
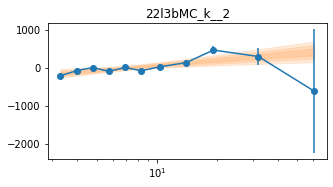
# Model the PHA data from fqlag mcmc with xspec; do the two frequencies simultaneously
os.chdir('%s/%s/lag/pha'%(base_dir, wdir))
for x in ['a', 'b', '']:
fit_pha_with_loglin('22l3bMC_k%s__1'%x, recalc=1, do_f2=1)
os.chdir('%s/%s'%(base_dir, wdir))
chains for 22l3bMC_ka__12
chains for 22l3bMC_kb__12
chains for 22l3bMC_k__12
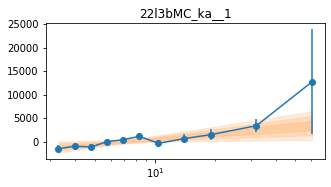
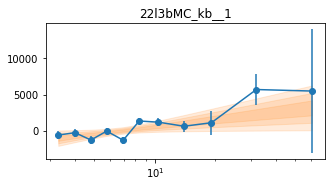
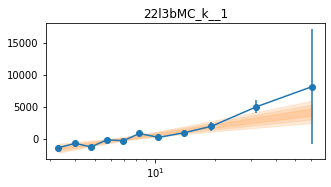
Different Frequency binning
Lag vs freq
Compare iron line to Compton hump and highest energies
# '3 3.3 3.6 4 4.4 4.8 5.2 5.7 6.3 6.9 7.6 8.3 9.1 10 11.7 13.8 16.2 19 22 31 42 58 79'
fqL, fqd = get_fq_bins(Lc[0], dt, mode=1, Nfq=5)
iEn = [[6,7,8], [16,17,18], [19,20,21]]
lag_fq_1 = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_fq_1.npz', iEn=iEn, iref=[0,1,2,3,4,5],
mcmc=[-4, 2000], logmod=False)
nfq: 7
fqL: 5.71089e-06 1.0782e-05 4.07122e-05 7.68634e-05 0.000145116 0.000273974 0.000517255 0.00195312
cache file lag/lag_fq_1.npz found. Reading ...!
# process mcmc from fqlag and plot histograms
lag_fqMC_1 = proc_lag_mcmc('lag_fq_1')
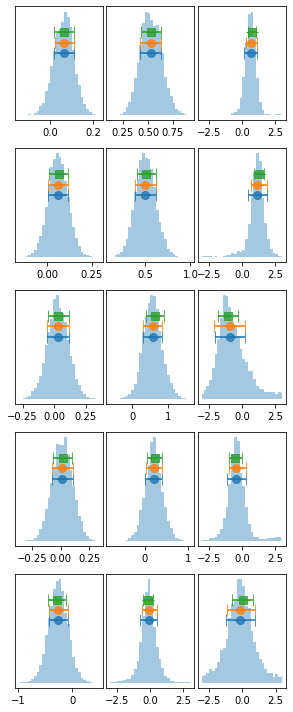
plot_lag(lag_fqMC_1)
write_lag(lag_fqMC_1, '_fqMC_1', pha=False, null_tests=False, pha_fq=True)
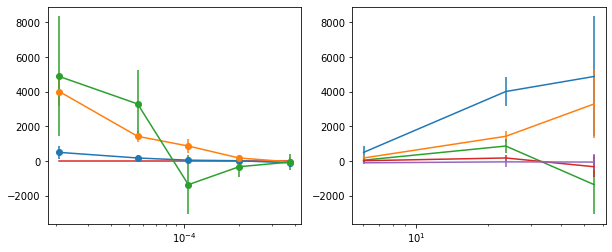
_e1: Followup the _fqMC__1 results
# 5.71089e-06 1.0782e-05 4.07122e-05 7.68634e-05 0.000145116 0.000273974 0.000517255 0.00195312
fqL = np.array([5e-6, 1e-5, 8e-5, 1.5e-4, 3e-4, 2e-3])
iEn = [[0,1], [2,3], [4,5], [6,7], [8,9], [10,11], [12,13], [14,15], [16,17], [18,19], [20,21]]
lag_e1 = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3b_e1.npz', iEn=iEn, mcmc=[-4, 2000], logmod=False)
cache file lag/lag_22l3b_e1.npz found. Reading ...!
# process mcmc from fqlag and plot histograms
lagMC_e1 = proc_lag_mcmc('lag_22l3b_e1')
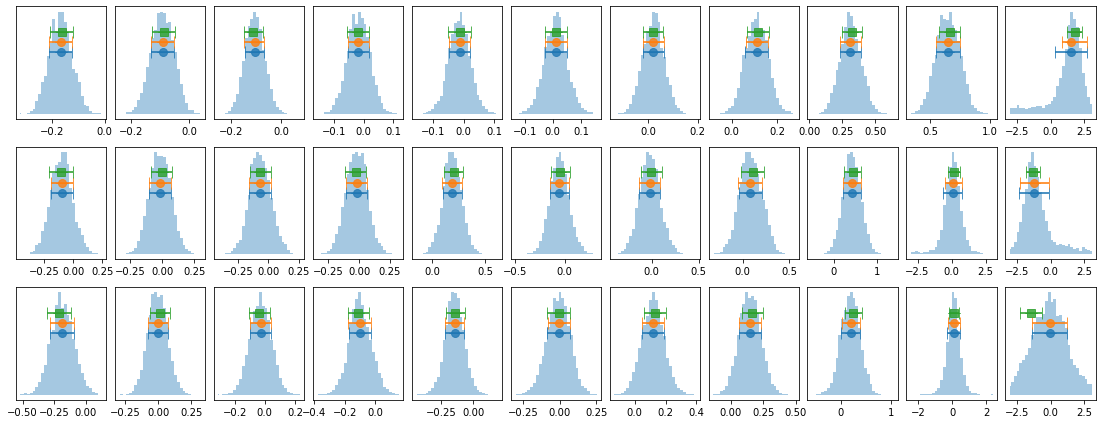
plot_lag(lagMC_e1)
write_lag(lagMC_e1, '_22l3bMC_e1', pha=True)
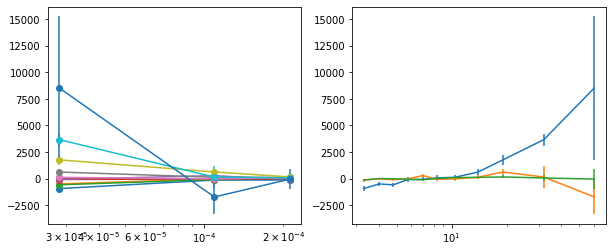
# Model the PHA data from fqlag mcmc with xspec
os.chdir('%s/%s/lag/pha'%(base_dir, wdir))
fit_pha_with_loglin('22l3bMC_e1__1', recalc=1)
fit_pha_with_loglin('22l3bMC_e1__2', recalc=1)
fit_pha_with_loglin('22l3bMC_e1__3', recalc=1)
os.chdir('%s/%s'%(base_dir, wdir))
chains for 22l3bMC_e1__1
chains for 22l3bMC_e1__2
chains for 22l3bMC_e1__3
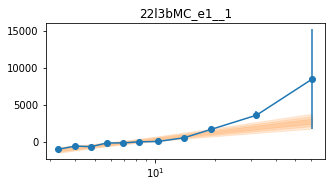
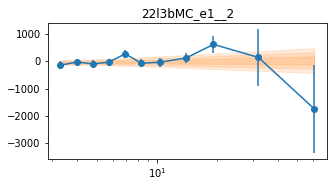
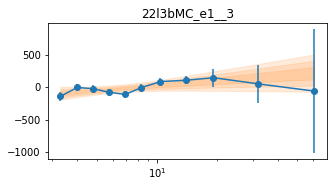
_e1 for individual groups
lag_e1a = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3b_e1a.npz', iEn=iEn, iLc=[0,1,2,3],
mcmc=[-4, 2000], logmod=False)
cache file lag/lag_22l3b_e1a.npz found. Reading ...!
lag_e1b = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3b_e1b.npz', iEn=iEn, iLc=[4,5,6,7,8],
mcmc=[-4, 2000], logmod=False)
cache file lag/lag_22l3b_e1b.npz found. Reading ...!
# process mcmc from fqlag and plot histograms
lagMC_e1 = proc_lag_mcmc('lag_22l3b_e1')
lagMC_e1a = proc_lag_mcmc('lag_22l3b_e1a')
lagMC_e1b = proc_lag_mcmc('lag_22l3b_e1b')
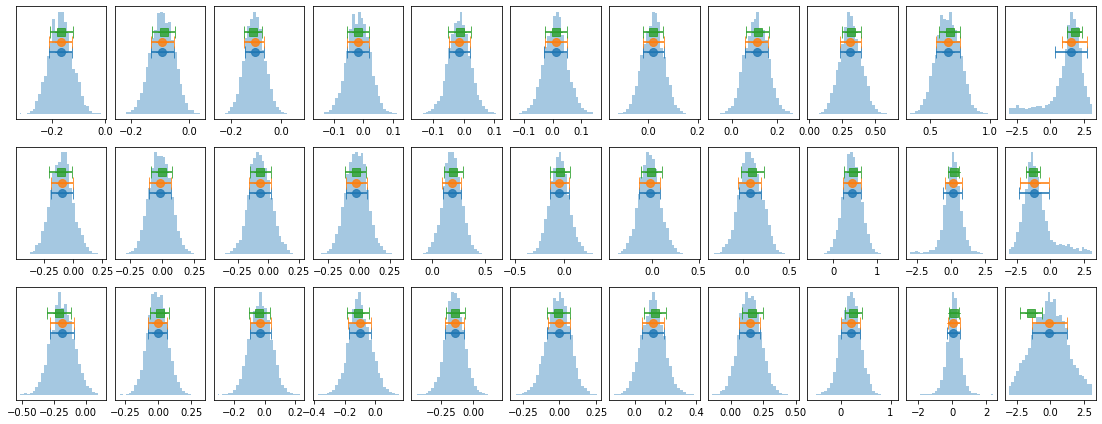
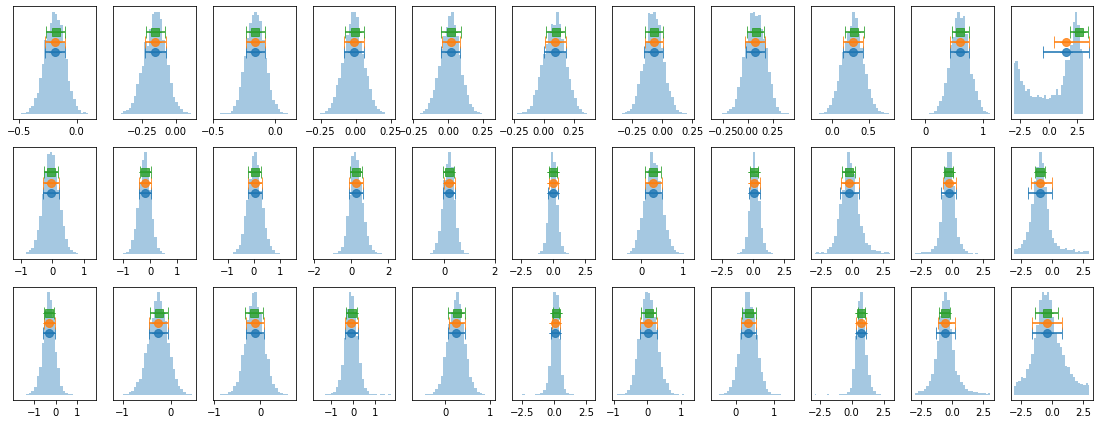
# plot lag-energy data from the mcmc chains #
for x in ['a', 'b', '']:
exec('plot_lag(lagMC_e1%s)'%x)
exec('write_lag(lagMC_e1%s, "_22l3bMC_e1%s", pha=True)'%(x,x))
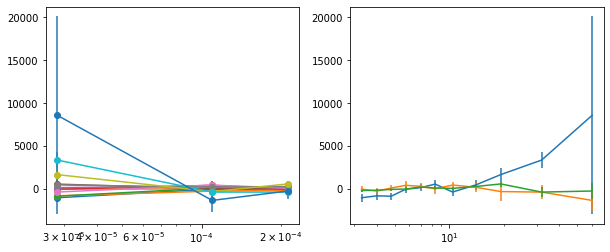
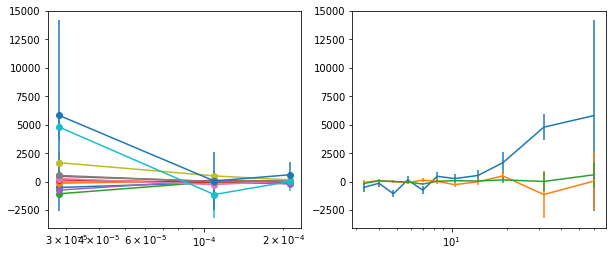
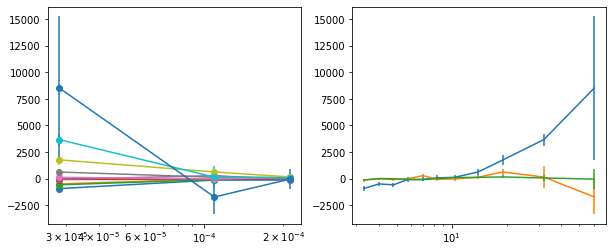
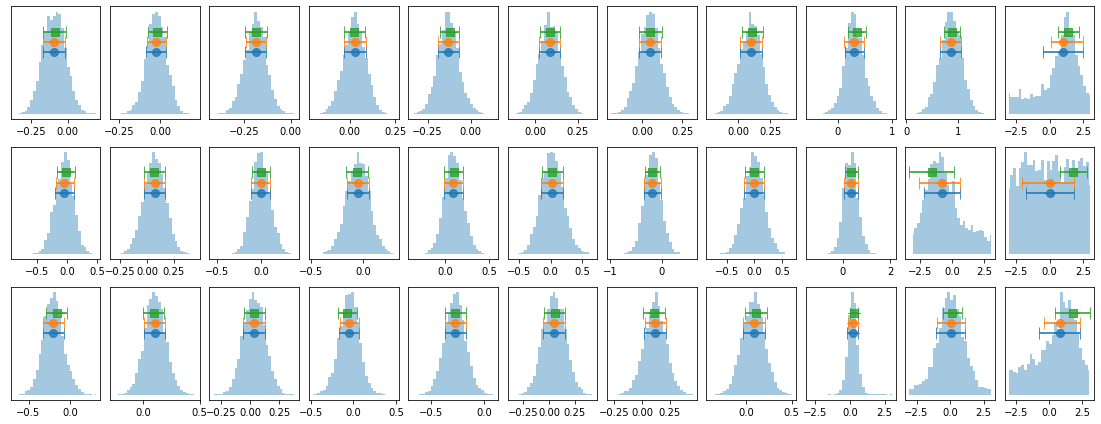
plot_lag(lagMC_e1)
_e1C: Similar _e1 but fit for the full cross spectrum instead of a transfer function
fqL = np.array([5e-6, 1e-5, 8e-5, 1.5e-4, 3e-4, 2e-3])
iEn = [[0,1], [2,3], [4,5], [6,7], [8,9], [10,11], [12,13], [14,15], [16,17], [18,19], [20,21]]
lag_e1C = calculate_lag_cross(Lc, fqL, dt, ebins, 'lag/lag_22l3b_e1C.npz', iEn=iEn, mcmc=[-4, 4000], logmod=False)
cache file lag/lag_22l3b_e1C.npz found. Reading ...!
# process mcmc from fqlag and plot histograms
lagMC_e1C = proc_lag_mcmc('lag_22l3b_e1C')
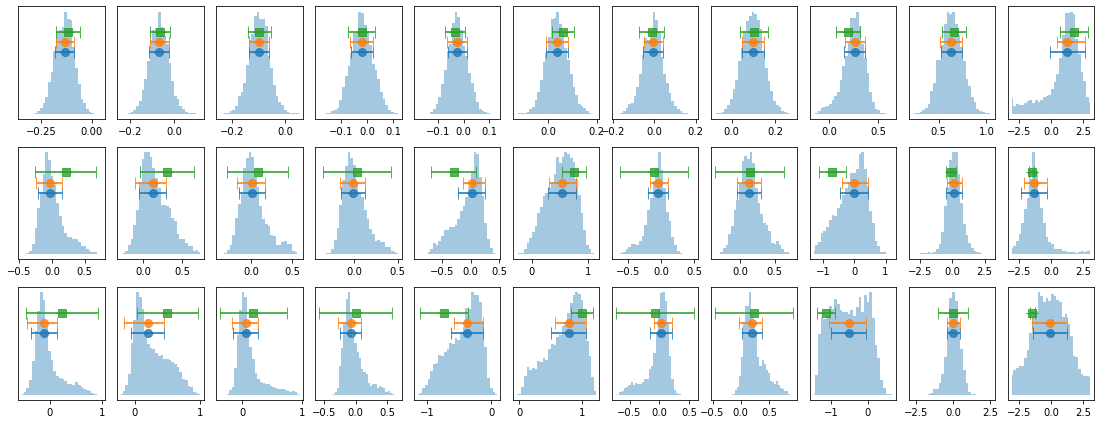
# write cross spectrum to pha files to model it
# (nen, 11, nfq); 11: fq, p,pe, amp,ampe, phi,phie(std), lag,lage, dphi-,dphi+
Res_mc, En, fqd, extra = lagMC_e1C
cxd,cxde = Res_mc[:,3], Res_mc[:,4]
os.chdir('lag/pha')
for i in [1,2,3]:
az.misc.write_pha_spec(En[0]-En[1], En[0]+En[1], cxd[:,i], cxde[:,i], f'cxd_22l3bMC_e1C__{i}')
os.chdir('../..')
cxd_22l3bMC_e1C__1.pha was created successfully
cxd_22l3bMC_e1C__2.pha was created successfully
cxd_22l3bMC_e1C__3.pha was created successfully
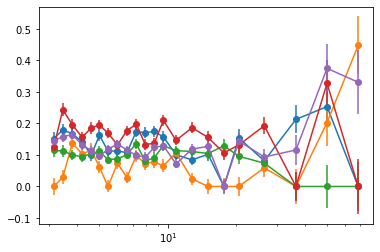
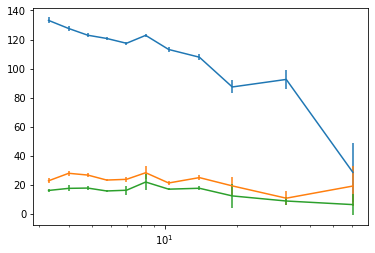
plt.errorbar(En[0], cxd[:,1], cxde[:,1])
plt.errorbar(En[0], cxd[:,2], cxde[:,2])
plt.errorbar(En[0], cxd[:,3], cxde[:,3])
plt.xscale('log')
#plt.yscale('log')
np.std?
# calculate the rms vs energy to estimate the variable spectrum #
s2 = np.array([[l[1].var(ddof=1) for l in lc] for lc in Lc])
e2 = np.array([[(l[2]**2).mean() for l in lc] for lc in Lc])
m2 = np.array([[l[1].mean()**2 for l in lc] for lc in Lc])
N = np.array([[len(l[1]) for l in lc] for lc in Lc])
fvar = (np.clip(s2 - e2, 0, np.inf)/m2)**0.5
fvarE = ((e2/N)/m2)**0.5
Fvar = fvar.mean(1)
FvarE = (np.sum(fvarE**2, 1)**0.5)/fvar.shape[1]
En = np.array(ebins.split(), np.double)
en = (En[1:] + En[:-1])/2
ene = (En[1:] - En[:-1])/2
for i in range(5):
plt.errorbar(en, fvar[:,i], fvarE[:,i], fmt='o-')
plt.xscale('log')