import os
import sys
base_dir = '/home/abzoghbi/data/mcg5-23-16/nustar_re_analysis'
if not base_dir in sys.path: sys.path.insert(0, base_dir)
from helpers import *
%load_ext autoreload
%autoreload 2
The autoreload extension is already loaded. To reload it, use:
%reload_ext autoreload
os.chdir(base_dir)
wdir = 'data/timing'
os.system('mkdir -p %s'%wdir)
os.chdir(wdir)
nu_obsids = np.array(['60001046002', '60001046004', '60001046006', '60001046008'])
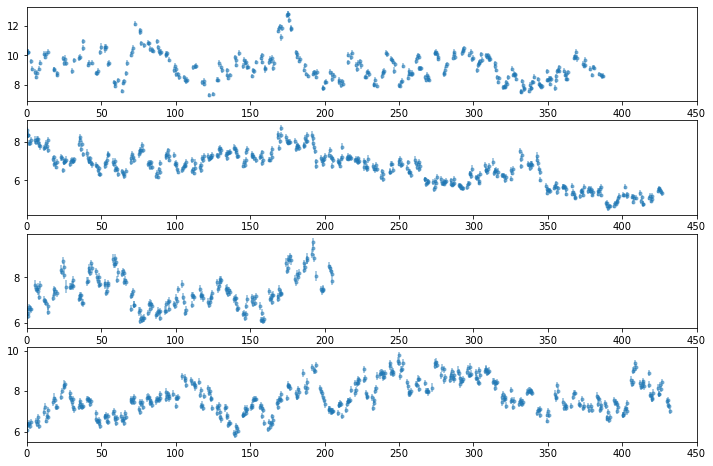
3-79 keV Total light curve
#!rm lc_1b_512.npz
loc_info, nen, dt = [base_dir, '1b'], 1, 512
LC = read_lc(loc_info, nu_obsids, dt, nen, combine_ab=True)
#!rm lc_22l3_512_bgd.npz
loc_info, nen, dt = [base_dir, '22l3'], 22, 512
LCb = read_lc(loc_info, nu_obsids, dt, nen, combine_ab=True, bgd=True)
LC = remove_high_bgd(LC, LCb)
reading data from lc_1b_512.npz ..
reading data from lc_22l3_512_bgd.npz ..
# plot light curves
nlc = len(LC[0])
fig, ax = plt.subplots(nlc, 1, figsize=(12, 8))
for ilc,lc in enumerate(LC[0]):
ax[ilc].errorbar((lc[0] - lc[0][0])/1e3, lc[1], lc[2], fmt='o', ms=3, alpha=0.5)
ax[ilc].set_xlim([0, 450])
dates = []
for o in nu_obsids:
with pyfits.open(f'{base_dir}/data/nustar/{o}_p/lc/1b/lc_512__a__1.lc') as fp:
dates.append(fp[1].header['date-obs'])
dates = TT.Time(dates)
# write lc
os.system('mkdir -p psd')
txt = ''
t0 = 0
for ilc,lc in enumerate(LC[0]):
tt = lc[0] - lc[0][0] + t0
t0 = tt[-1] + 1e4
tmjd = (dates[ilc] + TT.TimeDelta(lc[0] - lc[0][0], format='sec')).mjd
txt += '\ndescriptor t_%d rate_%d,+- tt_%d tmjd_%d\n'%(ilc+1,ilc+1,ilc+1, ilc+1)
txt += '\n'.join(['%10.10g %10.3g %10.3g %10.10g %10.7g'%(t,r,re,t_,_tm) for t,r,re,t_,_tm in
zip(lc[0]-lc[0][0], lc[1], lc[2], tt, tmjd)])
with open('psd/lc_%g.plot'%(dt), 'w') as fp: fp.write(txt)
PSD
# get segments #
tlen = 200
Lc, LcIdx = split_LC_to_segments(LC, tlen*1e3, plot=False)
# frequency bins
fqL, fqd = get_fq_bins(Lc[0], dt, mode=1, Nfq=10, nyquist=0.5)
psd_all_f1 = calculate_psd(Lc[0], fqL, 'psd/psd_all_f1.npz', errors=True)
nfq: 12
fqL: 2.31413e-06 3.61475e-06 1.12927e-05 1.76396e-05 2.75536e-05 4.30396e-05 6.72293e-05 0.000105014 0.000164036 0.00025623 0.000400239 0.000625187 0.00195312
# Another frequency binning
fqL, fqd = get_fq_bins(Lc[0], dt, mode=1, Nfq=16, nyquist=0.5)
psd_all_f2 = calculate_psd(Lc[0], fqL, 'psd/psd_all_f2.npz', errors=True)
nfq: 18
fqL: 2.31413e-06 3.1154e-06 8.38825e-06 1.12927e-05 1.52028e-05 2.04669e-05 2.75536e-05 3.70941e-05 4.99381e-05 6.72293e-05 9.05077e-05 0.000121846 0.000164036 0.000220834 0.000297299 0.000400239 0.000538823 0.000725393 0.00195312
-241.253 | 6.72 6.09 5.22 6.09 4.1 4.8 3.95 3.68 3.62 2.94 1.87 1.38 0.469 -0.256 -2.19 -4.19 -2.33 -4.39 | 1.32e-06
** done **
-241.253 | 6.72 6.09 5.22 6.09 4.1 4.8 3.95 3.68 3.62 2.94 1.87 1.38 0.469 -0.256 -2.19 -4.19 -2.33 -4.39 | 1.42e-06
** done **
## errors for param 0 ##
-241.253 -241.749 6.71822 7.53072 0.991894
## errors for param 1 ##
-241.253 -241.748 6.08609 6.68765 0.990162
## errors for param 2 ##
-241.253 -241.756 5.217 6.13888 1.00621
## errors for param 3 ##
-241.253 -241.749 6.08799 6.48643 0.991016
## errors for param 4 ##
-241.253 -241.753 4.09752 4.71081 0.99932
## errors for param 5 ##
-241.253 -241.749 4.80145 5.16082 0.991867
## errors for param 6 ##
-241.253 -241.755 3.94619 4.25869 1.00452
## errors for param 7 ##
-241.253 -241.749 3.67748 3.95091 0.991426
## errors for param 8 ##
-241.253 -241.749 3.61894 3.85722 0.992202
## errors for param 9 ##
-241.253 -241.75 2.94037 3.14545 0.993896
## errors for param 10 ##
-241.253 -241.751 1.86759 2.07657 0.995739
## errors for param 11 ##
-241.253 -241.758 1.3759 1.57316 1.00974
## errors for param 12 ##
-241.253 -241.756 0.46884 0.71884 1.00605
## errors for param 13 ##
-241.253 -241.75 -0.256087 0.0720384 0.99286
## errors for param 14 ##
-241.253 -241.752 -2.19453 -1.27265 0.997586
## errors for param 15 ##
-241.253 -241.751 -4.19081 -2.09121 0.994696
## errors for param 16 ##
-241.253 -241.752 -2.32877 -1.69205 0.997702
## errors for param 17 ##
-241.253 -241.751 -4.39141 -3.3543 0.995653
********************
6.72 6.09 5.22 6.09 4.1 4.8 3.95 3.68 3.62 2.94 1.87 1.38 0.469 -0.256 -2.19 -4.19 -2.33 -4.39
0.812 0.602 0.922 0.398 0.613 0.359 0.312 0.273 0.238 0.205 0.209 0.197 0.25 0.328 0.922 2.1 0.637 1.04
********************
# Analytical fit with PL and BPL
psd_all_f = calculate_psd(Lc[0], fqL, 'psd/psd_all_f.npz', do_bin=0, do_pl=1, do_bpl=1, errors=True)
-285.513 | -17.5 -2.1 | 7.72e-09
** done **
-285.513 | -17.5 -2.1 | 9.71e-06
** done **
## errors for param 0 ##
-285.513 -286.012 -17.5315 -17.0033 0.997351
## errors for param 1 ##
-285.513 -286.015 -2.09807 -2.04009 1.00242
********************
-17.5 -2.1
0.528 0.058
********************
-247.275 | -5.94 -3.3 -9.47 | 1.42e-07
** done **
-247.275 | -5.94 -3.3 -9.47 | 4.45e-07
** done **
## errors for param 0 ##
-247.275 -247.773 -5.94215 -5.79176 0.995061
## errors for param 1 ##
-247.275 -247.779 -3.29512 -3.05293 1.00798
## errors for param 2 ##
-247.275 -247.776 -9.46832 -9.32086 1.00245
********************
-5.94 -3.3 -9.47
0.15 0.242 0.147
********************
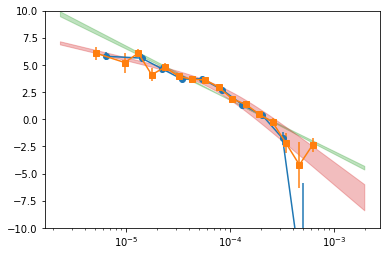
# plot the psd #
pBin = psd_all_f1['p_Bin']
plt.errorbar(pBin[0][1:-1], pBin[1][1:-1], pBin[2][1:-1], fmt='o-')
pBin = psd_all_f2['p_Bin']
plt.errorbar(pBin[0][1:-1], pBin[1][1:-1], pBin[2][1:-1], fmt='s-')
mPL = psd_all_f['m_pl']
plt.fill_between(mPL[0], mPL[1]-mPL[2], mPL[1]+mPL[2], alpha=0.3, color='C2')
mBPL = psd_all_f['m_bpl']
plt.fill_between(mBPL[0], mBPL[1]-mBPL[2], mBPL[1]+mBPL[2], alpha=0.3, color='C3')
plt.xscale('log')
plt.ylim([-10, 10])
(-10, 10)
# write psd_all #
txt = 'descriptor fq_f1 psd_f1,+-\n'
txt += '\n'.join(['%g %g %g'%(z[0], z[1], z[2]) for z in psd_all_f1['p_Bin'][:,1:-1].T])
txt += '\ndescriptor fq_f2 psd_f2,+-\n'
txt += '\n'.join(['%g %g %g'%(z[0], z[1], z[2]) for z in psd_all_f1['p_Bin'][:,1:-1].T])
txt += '\ndescriptor fq_pl psd_pl,+-\n'
txt += '\n'.join(['%g %g %g'%(z[0], z[1], z[2]) for z in psd_all_f['m_pl'].T])
txt += '\ndescriptor fq_bpl psd_bpl,+-\n'
txt += '\n'.join(['%g %g %g'%(z[0], z[1], z[2]) for z in psd_all_f['m_bpl'].T])
with open('psd/psd_all.plot', 'w') as fp: fp.write(txt)
p,pe = psd_all_f1['p_Bin'][1:,1:-1]
fqL = psd_all_f1['fqL'][1:-1]
az.misc.write_pha_spec(fqL[:-1], fqL[1:], p, pe, 'psd_f1')
p,pe = psd_all_f2['p_Bin'][1:,1:-1]
fqL = psd_all_f2['fqL'][1:-1]
az.misc.write_pha_spec(fqL[:-1], fqL[1:], p, pe, 'psd_f2')
os.system('mkdir -p psd/pha')
os.system('mv *pha *rsp *xsp psd/pha')
psd_f1.pha was created successfully
psd_f2.pha was created successfully
0