import os
import sys
base_dir = '/home/abzoghbi/data/mcg5-23-16/nustar_re_analysis'
if not base_dir in sys.path: sys.path.insert(0, base_dir)
from helpers import *
%load_ext autoreload
%autoreload 2
os.chdir(base_dir)
wdir = 'data/timing'
os.system('mkdir -p %s/lag'%wdir)
os.chdir(wdir)
nu_obsids = np.array(['60001046002', '60001046004', '60001046006', '60001046008'])
#!rm lc_22l3_512.npz
loc_info, nen, dt = [base_dir, '22l3'], 22, 512
LC = read_lc(loc_info, nu_obsids, dt, nen, combine_ab=True)
#!rm lc_22l3_512_bgd.npz
loc_info, nen, dt = [base_dir, '22l3'], 22, 512
LCb = read_lc(loc_info, nu_obsids, dt, nen, combine_ab=True, bgd=True)
LC = remove_high_bgd(LC, LCb)
reading data from lc_22l3_512.npz ..
reading data from lc_22l3_512_bgd.npz ..
# average counts per bin #
txt = '\n'.join([' '.join(['%8.4g'%(l[1].mean()*dt) for l in lc]) for lc in LC])
print(txt)
133.7 89.82 101.6 104.7
176 116.9 133.9 136.4
244.6 162 187.2 191.9
252.8 170.1 194.2 199.9
224.6 151.5 172.4 176.7
269.9 183.8 207.5 214.8
281.8 193.2 218.4 225.7
347.1 241.9 269.1 278.8
318.7 225.6 247.7 257.9
300.4 212.5 236.5 245.7
244 171.7 191.7 200.5
252 179.5 199.1 208.1
250.6 176.8 198.5 208
325.2 234.2 258.1 272.4
260.2 188.3 207.4 217.4
203.2 146.9 160.4 170
155.1 113.7 123.8 131.4
111.9 81.72 89.02 94.24
152.5 111.6 119.7 126.4
58.02 45.44 47.16 52.52
32.03 26.58 28.11 30.25
15.4 16.59 13.63 17.31
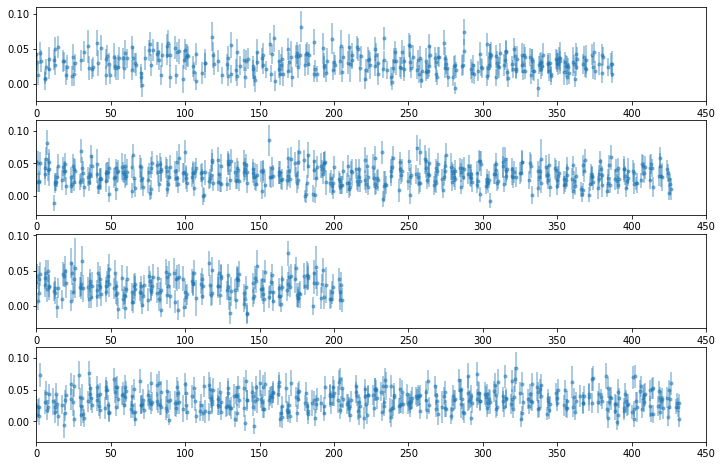
# plot light curves
ie = 21
nlc = len(LC[ie])
fig, ax = plt.subplots(nlc, 1, figsize=(12, 8))
for ilc,lc in enumerate(LC[ie]):
ax[ilc].errorbar((lc[0] - lc[0][0])/1e3, lc[1], lc[2], fmt='o', ms=3, alpha=0.5)
ax[ilc].set_xlim([0, 450])
Prepare Segments
ebins, dt = ('3 3.3 3.6 4 4.4 4.8 5.2 5.7 6.3 6.9 7.6 8.3 9.1 10 '
'11.7 13.8 16.2 19 22 31 42 58 79'), 512
tlen = 200
Lc, LcIdx = split_LC_to_segments(LC, tlen*1e3, plot=False)
_f7: Fig. 7 in Zoghbi+14: 6e-6 6e-4
Note that because of the segment lengths, the effective low bin bounday is 1e-5, so we use it
fqL = np.array([2e-6, 6e-6, 6e-4, 1.3e-3])
iEn = None
lag_f7 = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3_f7.npz', iEn=iEn, mcmc=[-4, 2000], logmod=False)
cache file lag/lag_22l3_f7.npz found. Reading ...!
lag_f7a = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3_f7a.npz', iEn=iEn, iLc=[0],
mcmc=[-4, 2000], logmod=False)
cache file lag/lag_22l3_f7a.npz found. Reading ...!
#lag_f7b = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3_f7b.npz', iEn=iEn, iLc=[1],
# mcmc=[-4, 2000], logmod=False)
#lag_f7c = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3_f7c.npz', iEn=iEn, iLc=[2],
# mcmc=[-4, 2000], logmod=False)
lag_f7d = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3_f7d.npz', iEn=iEn, iLc=[3],
mcmc=[-4, 2000], logmod=False)
cache file lag/lag_22l3_f7d.npz found. Reading ...!
lag_f7bc = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3_f7bc.npz', iEn=iEn, iLc=[1,2],
mcmc=[-4, 2000], logmod=False)
cache file lag/lag_22l3_f7bc.npz found. Reading ...!
# process mcmc from fqlag and plot histograms
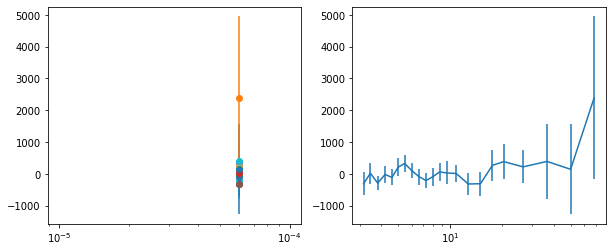
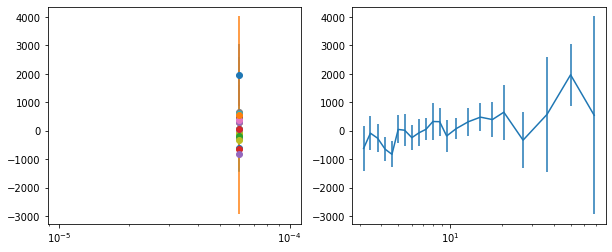
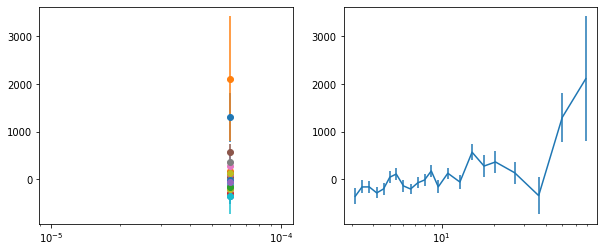
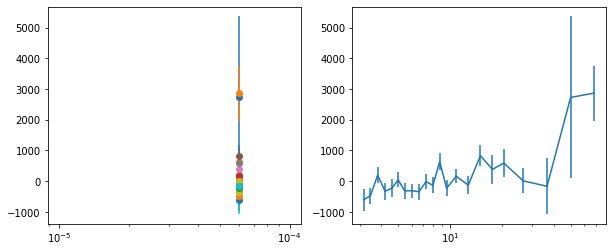
lagMC_f7 = proc_lag_mcmc('lag_22l3_f7')
lagMC_f7a = proc_lag_mcmc('lag_22l3_f7a')
lagMC_f7d = proc_lag_mcmc('lag_22l3_f7d')
lagMC_f7bc = proc_lag_mcmc('lag_22l3_f7bc')




for x in ['a', 'd', '', 'bc']:
exec('plot_lag(lagMC_f7%s)'%x)
exec('write_lag(lagMC_f7%s, "_22l3MC_f7%s", pha=True)'%(x,x))
### Model the PHA data with xspec
os.chdir('%s/%s/lag/pha'%(base_dir, wdir))
for x in ['a', 'bc', 'd', '']:
fit_pha_with_loglin('22l3MC_f7%s__1'%x, recalc=1)
os.chdir('%s/%s'%(base_dir, wdir))
chains for 22l3MC_f7a__1
chains for 22l3MC_f7bc__1
chains for 22l3MC_f7d__1
chains for 22l3MC_f7__1
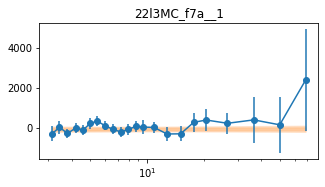
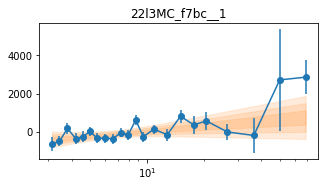
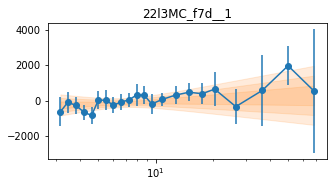
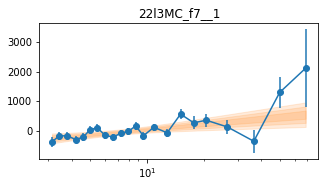
Followup the _fqMC__1 results (in lag_22l3b)
# 2.31413e-06 4.97082e-06 2.13549e-05 4.5871e-05 9.85324e-05 0.000211651 0.000454632 0.00195312
fqL = np.array([2e-6, 5e-6, 2e-5, 1e-4, 2e-4, 2e-3])
iEn = None
lag_e1 = calculate_lag(Lc, fqL, dt, ebins, 'lag/lag_22l3_e1.npz', iEn=iEn, mcmc=[-4, 2000], logmod=False)
cache file lag/lag_22l3_e1.npz found. Reading ...!
# process mcmc from fqlag and plot histograms
lagMC_e1 = proc_lag_mcmc('lag_22l3_e1')
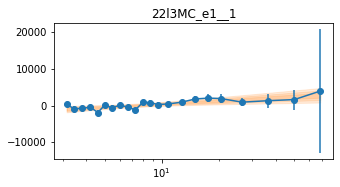
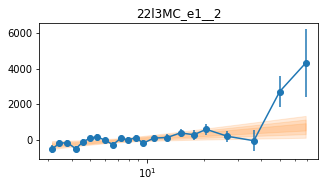
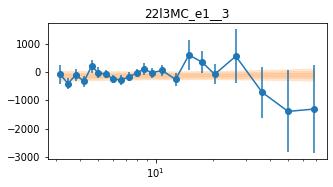
plot_lag(lagMC_e1)
write_lag(lagMC_e1, '_22l3MC_e1', pha=True)
# Model the PHA data from fqlag mcmc with xspec
os.chdir('%s/%s/lag/pha'%(base_dir, wdir))
fit_pha_with_loglin('22l3MC_e1__1', recalc=1)
fit_pha_with_loglin('22l3MC_e1__2', recalc=1)
fit_pha_with_loglin('22l3MC_e1__3', recalc=1)
os.chdir('%s/%s'%(base_dir, wdir))
chains for 22l3MC_e1__1
chains for 22l3MC_e1__2
chains for 22l3MC_e1__3