Description
- Move the spectra from individual observation folder to one folder
- Correct for the gain in the pn data
- Move the spectra from sub-segments and correct their gains. Assuming a single gain correction per observation.
- Move the Suzaku and NuSTAR spectra to
xmm_spec
import sys,os
base_dir = '/u/home/abzoghbi/data/ngc4151/spec_analysis'
sys.path.append(base_dir)
from spec_helpers import *
%load_ext autoreload
%autoreload 2
Read useful data from data notebook
# define and read some useful descriptive information #
data_dir = 'data/xmm'
spec_dir = 'data/xmm_spec'
os.chdir('%s/%s'%(base_dir, data_dir))
data = np.load('log/data.npz')
spec_obsids = data['spec_obsids']
obsids = data['obsids']
spec_data = data['spec_data']
spec_ids = [i+1 for i,o in enumerate(obsids) if o in spec_obsids]
Move the spectra to one location
os.chdir(base_dir)
if not os.path.exists(spec_dir):
os.system('mkdir -p %s'%spec_dir)
os.chdir(spec_dir)
for o in spec_obsids:
os.system('cp %s/%s/%s/pn/spec/spec_* .'%(base_dir, data_dir, o))
os.system('cp %s/%s/%s/mos/spec/spec_* .'%(base_dir, data_dir, o))
os.system('cp %s/%s/%s/rgs/spec_rgs* .'%(base_dir, data_dir, o))
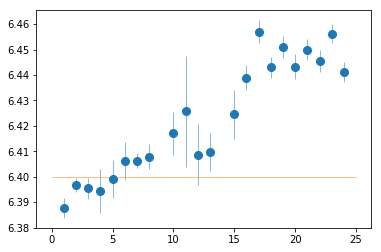
The energy of the FeK line
PN spectra
- Fitting is done with
xspec - It uses
fit_1infit.tcl
os.chdir('%s/%s'%(base_dir, spec_dir))
os.system('mkdir -p fits')
fit_1 = fit_xspec_model('fit_1', spec_ids, base_dir, '_pn')
plt.errorbar(spec_ids, fit_1[:,2,0], fit_1[:,2,1], fmt='o', ms=8, lw=0.5)
plt.plot([0,25], [6.4]*2, lw=0.5)
[<matplotlib.lines.Line2D at 0x7f84200c3a20>]
MOS Spectra
do mos-2 only, as mos-1 has some missing spectra, and the gain appear to be wrong too.
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_1_m2 = fit_xspec_model('fit_1', spec_ids, base_dir, '_m2', spec_root='spec_m2_%d.grp')
plt.errorbar(spec_ids, fit_1[:,2,0], fit_1[:,2,1], fmt='o', ms=8, lw=0.5)
plt.errorbar(spec_ids, fit_1_m2[:,2,0], fit_1_m2[:,2,1], fmt='s', ms=8, lw=0.5)
plt.plot([0,25], [6.4]*2, lw=0.5)
plt.xlabel('Obs num'); _=plt.ylabel('Line Energy (keV)')
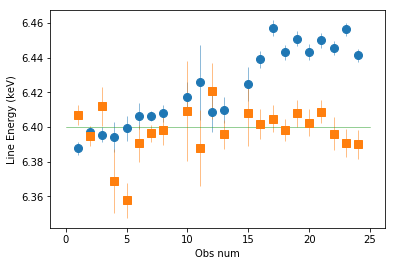
The mos-2 data is more consistent with 6.4. The PN is affected by gain
# save some of these results #
os.system('mkdir -p results/prepare')
text = 'descriptor iobs id en_pn,+- en_m2,+-\n'
text += '\n'.join(['{:5} {:5} {:8.4} {:8.4} {:8.4} {:8.4}'.format(i, spec_ids[i],
fit_1[i,2,0], fit_1[i,2,1], fit_1_m2[i,2,0], fit_1_m2[i,2,1])
for i in range(len(fit_1))])
#print(text)
with open('results/prepare/line_en.plot', 'w') as fp: fp.write(text)
Track the soft lines too
os.chdir('%s/%s'%(base_dir, spec_dir))
soft_spec_pn = fit_xspec_model('fit_1a', spec_ids, base_dir, '_pn')
soft_spec_m2 = fit_xspec_model('fit_1a', spec_ids, base_dir, '_m2', spec_root='spec_m2_%d.grp')
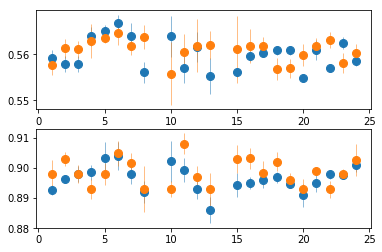
plt.subplot(211)
plt.errorbar(spec_ids, soft_spec_pn[:,2,0], soft_spec_pn[:,2,1], fmt='o', ms=8, lw=0.5)
plt.errorbar(spec_ids, soft_spec_m2[:,2,0], soft_spec_m2[:,2,1], fmt='o', ms=8, lw=0.5)
plt.subplot(212)
plt.errorbar(spec_ids, soft_spec_pn[:,4,0], soft_spec_pn[:,4,1], fmt='o', ms=8, lw=0.5)
plt.errorbar(spec_ids, soft_spec_m2[:,4,0], soft_spec_m2[:,4,1], fmt='o', ms=8, lw=0.5)
<ErrorbarContainer object of 3 artists>
# save some of these results #
text = '\ndescriptor en_pn_s1,+- en_pn_s2,+- en_m2_s1,+- en_m2_s2,+-\n'
text += '\n'.join(['{:8.4} {:8.4} {:8.4} {:8.4} {:8.4} {:8.4} {:8.4} {:8.4}'.format(
soft_spec_pn[i,2,0], soft_spec_pn[i,2,1], soft_spec_pn[i,4,0], soft_spec_pn[i,4,1],
soft_spec_m2[i,2,0], soft_spec_m2[i,2,1], soft_spec_m2[i,4,0], soft_spec_m2[i,4,1])
for i in range(len(soft_spec_pn))])
#print(text)
with open('results/prepare/line_en.plot', 'a') as fp: fp.write(text)
PN and MOS are consistent in the soft band, hence gain correction is needed
Finding Gain Correction
We use linear function that corrects the energy of the FeK line and leaves the soft energies (we use 0.9 keV) unchanged.
# soft_en = 0.9
# slopes = (pn_spec[:,2,0] - soft_en) / (6.4 - soft_en)
# offsets = soft_en - soft_en*slopes
# plt.subplot(211); plt.plot(spec_ids, slopes, 'o-')
# plt.subplot(212); plt.plot(spec_ids, offsets, 'o-')
Regroup RGS spectra
os.chdir('%s/%s'%(base_dir, spec_dir))
for ispec in spec_ids:
cmd = 'export HEADASNOQUERY=;export HEADASPROMPT=/dev/null;'
cmd += ('ftgrouppha spec_rgs_{0}.grp spec_rgs_{0}.g snmin 6').format(ispec)
p = subp.call(['/bin/bash', '-i', '-c', cmd])
Fit the RGS spectrra: fit_gain_1
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_xspec_model('fit_gain_1', spec_ids, base_dir, spec_root='spec_rgs_%d.g', read_fit=False)
Use rgs model in PN to obtain the gain: fit_gain_2
# run in serial, because xspec steppar is run in parallel.
os.chdir('%s/%s'%(base_dir, spec_dir))
for ispec in spec_ids:
if not os.path.exists('fits/fit_gain_2g__%s.xcm'%ispec):
tcl = 'source %s/fit.tcl\n'%base_dir
tcl += 'fit_gain_2 %d\nexit\n'%(ispec)
xcm = 'tmp_%d.xcm'%ispec
with open(xcm, 'w') as fp: fp.write(tcl)
cmd = 'xspec - %s > tmp_%d.log 2>&1'%(xcm, ispec)
p = subp.call(['/bin/bash', '-i', '-c', cmd])
# read the gains #
gdata = np.array([np.loadtxt('fits/fit_gain_2g__%d.log'%i) for i in spec_ids])
slopes = gdata[:,-2]
offsets = gdata[:,-1]
plt.subplot(211); plt.plot(spec_ids, slopes, 'o-')
plt.subplot(212); plt.plot(spec_ids, offsets, 'o-')
[<matplotlib.lines.Line2D at 0x7f84b3db8cf8>]
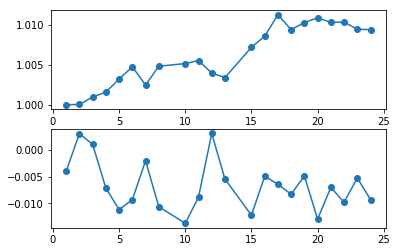
Apply the gain to the response files
os.chdir('%s/%s'%(base_dir, spec_dir))
print('{:5} {:12} {:10} {:10} {:10}'.format('num', 'obsid', 'slope', 'offset', 'apply?'))
for ii,ispec in enumerate(spec_ids):
#d = np.loadtxt('fits/fit_gain_pn__%dg.log'%ispec)
slope, offset, apply = slopes[ii], offsets[ii], gdata[ii,-3] < 1
text = '{:5} {:12} {:10.4} {:10.4} {:10}'.format(
ispec, spec_obsids[ii], slope, offset, apply)
print(text)
#continue
# apply gain #
if not apply: continue
rmf = 'spec_%d.rmf'%(ispec)
arf = 'spec_%d.arf'%(ispec)
# rmf #
with pyfits.open(rmf) as fp:
if not 'GAINPARS' in fp[1].header.keys():
fp[1].data['energ_lo'] = fp[1].data['energ_lo']/slope - offset
fp[1].data['energ_hi'] = fp[1].data['energ_hi']/slope - offset
fp[2].data['e_min'] = fp[2].data['e_min']/slope - offset
fp[2].data['e_max'] = fp[2].data['e_max']/slope - offset
fp[1].header['GAINPARS'] = '{}, {}'.format(slope, offset)
os.system('rm %s.shift >/dev/null 2>&1'%rmf)
fp.writeto('%s.shift'%rmf)
os.system('mv %s %s.off'%(rmf, rmf))
os.system('mv %s.shift %s'%(rmf, rmf))
# arf #
with pyfits.open(arf) as fp:
if not 'GAINPARS' in fp[1].header.keys():
fp[1].data['energ_lo'] = fp[1].data['energ_lo']/slope - offset
fp[1].data['energ_hi'] = fp[1].data['energ_hi']/slope - offset
fp[1].header['GAINPARS'] = '{}, {}'.format(slope, offset)
os.system('rm %s.shift >/dev/null 2>&1'%arf)
fp.writeto('%s.shift'%arf)
os.system('mv %s %s.off'%(arf, arf))
os.system('mv %s.shift %s'%(arf, arf))
num obsid slope offset apply?
1 0112310101 1.0 -0.004 1
2 0112830201 1.0 0.003093 1
3 0112830501 1.001 0.001058 1
4 0143500101 1.002 -0.007168 1
5 0143500201 1.003 -0.0112 1
6 0143500301 1.005 -0.00936 1
7 0402660101 1.002 -0.002021 1
8 0402660201 1.005 -0.01068 1
10 0657840301 1.005 -0.01379 1
11 0657840401 1.006 -0.008874 1
12 0679780101 1.004 0.00312 1
13 0679780201 1.003 -0.005462 1
15 0679780401 1.007 -0.01226 1
16 0761670101 1.009 -0.00496 1
17 0761670201 1.011 -0.006429 1
18 0761670301 1.009 -0.008327 1
19 0761670401 1.01 -0.004856 1
20 0761670501 1.011 -0.01301 1
21 0761670601 1.01 -0.00703 1
22 0761670701 1.01 -0.009841 1
23 0761670801 1.009 -0.005254 1
24 0761670901 1.009 -0.009328 1
# group spectra optimally using ftgrouppha
os.chdir('%s/%s'%(base_dir, spec_dir))
for ispec in spec_ids:
cmd = 'export HEADASNOQUERY=;export HEADASPROMPT=/dev/null;'
cmd += 'ftgrouppha spec_{0}.grp tmp_{0}.grp opt respfile=spec_{0}.rmf;'.format(ispec)
cmd += 'mv tmp_{0}.grp spec_{0}.grp'.format(ispec)
_ = os.system(cmd)
SubSpec
### Read useful data from data notebook
data_dir = 'data/xmm'
subspec_dir = 'data/xmm_subspec'
os.chdir('%s/%s'%(base_dir, data_dir))
data = np.load('log/data.npz')
iselect = data['tselect_ispec']
Move the files to one location
### Read useful data from data notebook
os.system('mkdir -p %s/%s'%(base_dir, subspec_dir))
os.chdir('%s/%s'%(base_dir, subspec_dir))
os.system('for i in `find ../xmm|grep subspec$`;do cp $i/spec* .;done')
0
Apply gain
os.chdir('%s/%s'%(base_dir, subspec_dir))
for ii,idx in enumerate(spec_ids):
slope, offset, apply = slopes[ii], offsets[ii], True
# apply gain #
for ispec in iselect[ii]:
rmf = 'spec_%d.rmf'%(ispec)
arf = 'spec_%d.arf'%(ispec)
# rmf #
with pyfits.open(rmf) as fp:
if not 'GAINPARS' in fp[1].header.keys():
fp[1].data['energ_lo'] = fp[1].data['energ_lo']/slope - offset
fp[1].data['energ_hi'] = fp[1].data['energ_hi']/slope - offset
fp[2].data['e_min'] = fp[2].data['e_min']/slope - offset
fp[2].data['e_max'] = fp[2].data['e_max']/slope - offset
fp[1].header['GAINPARS'] = '{}, {}'.format(slope, offset)
os.system('rm %s.shift >/dev/null 2>&1'%rmf)
fp.writeto('%s.shift'%rmf)
os.system('mv %s %s.off'%(rmf, rmf))
os.system('mv %s.shift %s'%(rmf, rmf))
# arf #
with pyfits.open(arf) as fp:
if not 'GAINPARS' in fp[1].header.keys():
fp[1].data['energ_lo'] = fp[1].data['energ_lo']/slope - offset
fp[1].data['energ_hi'] = fp[1].data['energ_hi']/slope - offset
fp[1].header['GAINPARS'] = '{}, {}'.format(slope, offset)
os.system('rm %s.shift >/dev/null 2>&1'%arf)
fp.writeto('%s.shift'%arf)
os.system('mv %s %s.off'%(arf, arf))
os.system('mv %s.shift %s'%(arf, arf))
Move Suzaku & NuSTAR data to the same location too
suz_dir = 'data/suzaku'
nu_dir = 'data/nustar'
os.chdir('%s/%s'%(base_dir, spec_dir))
for o in glob.glob('%s/%s/*_p'%(base_dir, suz_dir)):
os.system('cp %s/spec/spec*fi* .'%o)
for o in glob.glob('%s/%s/*_p'%(base_dir, nu_dir)):
os.system('cp %s/spec/spec*.??? .'%o)