Content
- Unfolded spectra
- Exploring the hard band:
- Fe K edge energy:
fit_2(6-10 keV). - Fe K edge: Neutral vs ionized:
fit_2a,fit_2b. - Extend fit of neutral absorption down to 4 keV,
fit_2c. - Add high xi absorber,
fit_2d - Curvature in residuals, model with:
- Low xi WA,
fit_2e - Broad gausian,
fit_2f
- Low xi WA,
- Extend analysis to 2.5 keV,
fit_2g - Add a powerlaw (PC/Scattering),
fit_2h- Test a WA instead of the extra powerlaw,
fit_2h1
- Test a WA instead of the extra powerlaw,
- Use xillver instead of lines for K$\alpha$ & K$\beta$,
fit_2i, fromfit_2h- Test again a WA instead of the extra powerlaw,
fit_2i1
- Test again a WA instead of the extra powerlaw,
- Fe K edge energy:
- Broadband Characterization of the Spectra
- Soft variability: Evidence for Warm absorber
- Hard Spectrum: The need for Partial Covering
import sys,os
base_dir = '/u/home/abzoghbi/data/ngc4151/spec_analysis'
sys.path.append(base_dir)
from spec_helpers import *
%load_ext autoreload
%autoreload 2
### Read useful data from data notebook
data_dir = 'data/xmm'
spec_dir = 'data/xmm_spec'
os.chdir('%s/%s'%(base_dir, data_dir))
data = np.load('log/data.npz')
spec_obsids = data['spec_obsids']
obsids = data['obsids']
spec_data = data['spec_data']
spec_ids = [i+1 for i,o in enumerate(obsids) if o in spec_obsids]
Plot The spectra
- plot the unfolded spectra from all the 22 datasets.
os.chdir('%s/%s'%(base_dir, spec_dir))
os.system('mkdir -p results/explore')
nspec = len(spec_obsids)
tcl = 'source %s/fit.tcl\nsetpl ener\n'%base_dir
for ispec in spec_ids:
tcl += ('da spec_%d.grp\n'
'ign 0.0-.3,10.-**\n'
'az_plot_unfold u tmp_%d pn_%d\n')%((ispec,)*3)
with open('tmp.xcm', 'w') as fp: fp.write(tcl)
cmd = 'xspec - tmp.xcm > tmp.log 2>&1'
subp.call(['/bin/bash', '-i', '-c', cmd])
os.system('cat tmp_*plot > results/explore/spec_unfold.plot')
os.system('rm tmp_*plot tmp.???')
0
fit_2: Iron Edge at 7 keV: Model with zedge
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_2 = fit_xspec_model('fit_2', spec_ids, base_dir)
# plot edge energy and tau #
fig = plt.figure(figsize=(8,4))
ax = plt.subplot(221)
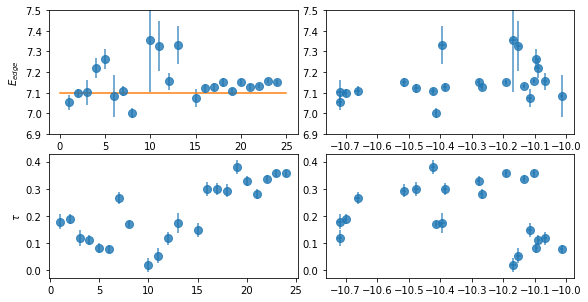
plt.errorbar(spec_ids, fit_2[:,0,0], fit_2[:,0,1], fmt='o', ms=8, alpha=0.8)
ax.set_ylabel(r'$E_{edge}$')
plt.plot([0,25], [7.1]*2)
ax.set_ylim([6.9,7.5])
ax = plt.subplot(223)
plt.errorbar(spec_ids, fit_2[:,1,0], fit_2[:,1,1], fmt='o', ms=8, alpha=0.8)
ax.set_ylabel(r'$\tau$')
ax = plt.subplot(222)
ax.set_ylim([6.9,7.5])
plt.errorbar(fit_2[:,2,0], fit_2[:,0,0], fit_2[:,0,1], xerr=fit_2[:,2,1], fmt='o', ms=8, alpha=0.8)
ax = plt.subplot(224)
plt.errorbar(fit_2[:,2,0], fit_2[:,1,0], fit_2[:,1,1], xerr=fit_2[:,2,1], fmt='o', ms=8, alpha=0.8)
plt.tight_layout(pad=0)
bins = np.linspace(6.9, 7.5, 10)
hist_2 = plt.hist(fit_2[:,0,0], bins)
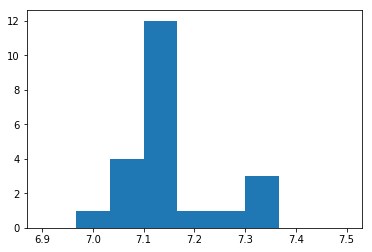
The edge energy is consistent with neutral except for a few cases where the enrgy is ~7.3, suggesting some ionized absorption
# write the energy values and histogram #
text = 'descriptor iobs edge_e,+,- pflx,+,-\n'
text += '\n'.join(['{} {} {} {} {} {} {}'.format(*z) for z in zip(spec_ids,
fit_2[:,0,0], fit_2[:,0,2], -fit_2[:,0,3], fit_2[:,2,0], fit_2[:,2,2], -fit_2[:,2,3])])
#print(text)
text += '\ndescriptor h_x,+- h_y\n'
text += '\n'.join(['{} {} {}'.format(*z) for z in zip(
(hist_2[1][1:]+hist_2[1][:-1])/2, (hist_2[1][1:]-hist_2[1][:-1])/2, hist_2[0])])
with open('results/explore/edge_en_hist.plot', 'w') as fp: fp.write(text)
fit_2a/fit_2b: Iron Edge at 7 keV: Neutral vs ionized
Compare ztbabs to zxipcf
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_xspec_model('fit_2ab', spec_ids, base_dir, read_fit=0)
# check the fit results; we need only the stats #
fit_2_n = np.array([open('fits/fit_2a__%d.log'%ispec).readlines()[1].split()[1:]
for ispec in spec_ids], np.double)
fit_2_x = np.array([open('fits/fit_2b__%d.log'%ispec).readlines()[1].split()[1:]
for ispec in spec_ids], np.double)
f_indiv = ftest(fit_2_x[:,0], fit_2_x[:,1], fit_2_n[:,0], fit_2_n[:,1])
f_tot = ftest(fit_2_x[:,0].sum(), fit_2_x[:,1].sum(), fit_2_n[:,0].sum(), fit_2_n[:,1].sum())
fmt = '{:8} {:8.5} {:6} {:8.5} {:6} {:8.5} {:8.5}'
text = fmt.format('id', 'c2_1', 'nd_1', 'c2_2', 'nd_2', 'fstat', 'fpval') + '\n'
text += '\n'.join([fmt.format(spec_ids[i],
fit_2_n[i,0], fit_2_n[i,1], fit_2_x[i,0], fit_2_x[i,1], f_indiv[0][i], f_indiv[2][i])
for i in range(nspec)])
text += '\n' + fmt.format('tot', fit_2_n[:,0].sum(), fit_2_n[:,1].sum(), fit_2_x[:,0].sum(),
fit_2_x[:,1].sum(), f_tot[0], f_tot[2])
print(text)
id c2_1 nd_1 c2_2 nd_2 fstat fpval
1 39.776 32.0 38.983 31.0 0.63109 0.43299
2 42.275 32.0 46.771 31.0 -2.9804 1.0
3 35.349 29.0 36.661 28.0 -1.0017 1.0
4 46.81 34.0 36.691 33.0 9.1007 0.0048918
5 45.121 34.0 39.061 33.0 5.1198 0.030361
6 72.595 34.0 57.052 33.0 8.9906 0.0051278
7 43.768 30.0 59.04 29.0 -7.5012 1.0
8 73.714 31.0 66.445 30.0 3.282 0.080068
10 21.512 27.0 21.274 26.0 0.29136 0.59394
11 28.828 28.0 26.062 27.0 2.8657 0.102
12 49.198 28.0 31.032 27.0 15.805 0.00047212
13 37.175 27.0 29.015 26.0 7.3119 0.011917
15 51.398 28.0 47.517 27.0 2.2051 0.14913
16 42.163 28.0 48.015 27.0 -3.2907 1.0
17 31.878 28.0 53.876 27.0 -11.024 1.0
18 34.723 28.0 42.31 27.0 -4.8414 1.0
19 36.986 28.0 63.343 27.0 -11.235 1.0
20 36.511 29.0 57.889 28.0 -10.34 1.0
21 44.092 30.0 80.577 29.0 -13.131 1.0
22 63.807 30.0 109.4 29.0 -12.086 1.0
23 138.86 30.0 179.4 29.0 -6.5529 1.0
24 100.84 30.0 159.65 29.0 -10.682 1.0
tot 1117.4 655.0 1330.1 633.0 -4.6008 1.0
## plot the residuals for the neutral vs ionized absorber ##
extra_cmd ='del 2; del 3; del 4; freez [az_free]; thaw 4 8; new 12=0.12*8;fit'
write_resid(base_dir, spec_ids, '2a', extra_cmd, -1, avg_bin=False, outdir='results/explore')
extra_cmd ='del 2; del 3; del 4; freez [az_free]; thaw 6 10; new 14=0.12*10;fit'
write_resid(base_dir, spec_ids, '2b', extra_cmd, -1, avg_bin=False, outdir='results/explore')
Neutral absorber gives superior fit in about half the cases. In the other half, the improvement is only marginal (pvalue > 0.0027), except for obs-12 where p=0.00047
The spectra in fit_2a,2b rise at the lower boundary (6 keV), so we extend it to 4 keV
fit_2c: Extend Nuetral absorber (fit_2a) to 4 keV
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_2c = fit_xspec_model('fit_2c', spec_ids, base_dir)
# plot the result #
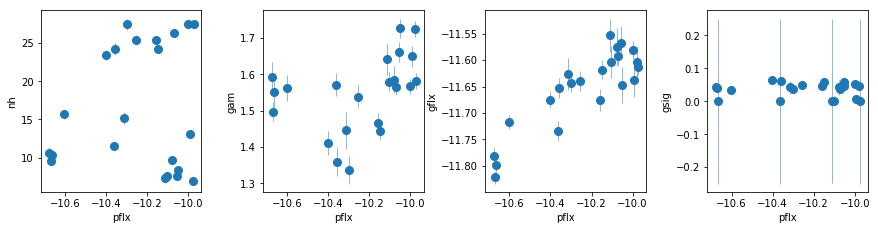
par_names = ['nh', 'pflx', 'gam', 'gflx', 'gsig']
fit = fit_2c
fig = plt.figure(figsize=(12,3))
idx = [0,2,3,4]; iref = 1
for i,ix in enumerate(idx):
ax = plt.subplot(1,len(idx),i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
# write the residuals #
extra_cmd ='del 2; del 3; del 4; freez [az_free]; thaw 4 8; new 12=0.12*8;fit'
write_resid(base_dir, spec_ids, '2c', extra_cmd, -1, avg_bin=True, outdir='results/explore')
The fit is does a good job for the low flux observations. For high fluxes, there an absorption line that is stronger for higher flux at ~6.7 keV, that is clear in the total residuals too
fit_2d: Add a high $\xi$ absorberon top of the neutral
We use zxipcf (standard XSTAR table)
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_2d = fit_xspec_model('fit_2d', spec_ids, base_dir)
# plot the result #
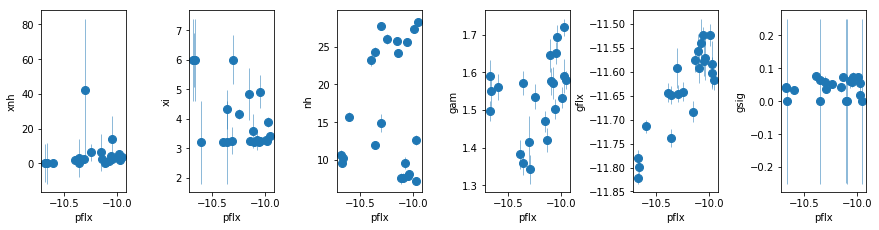
par_names = ['xnh', 'xi', 'nh', 'pflx', 'gam', 'gflx', 'gsig']
fit = fit_2d
fig = plt.figure(figsize=(12,3))
idx = [0,1,2,4,5,6]; iref = 3
for i,ix in enumerate(idx):
ax = plt.subplot(1,len(idx),i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
# write the residuals #
extra_cmd ='del 3; del 4; del 5; freez [az_free]; thaw 8 12; new 16=0.12*12;fit'
write_resid(base_dir, spec_ids, '2d', extra_cmd, -1, avg_bin=True, outdir='results/explore')
It appears like the high ionization absorber is present at high fluxes only. Investigate in detail later This model describes the data resonably well (p<0.002) except for two cases, 2, 8, 23, and 24. The average residuals show a broad curvature. We test two models: a low-xi WA or a broad gaussian emission line
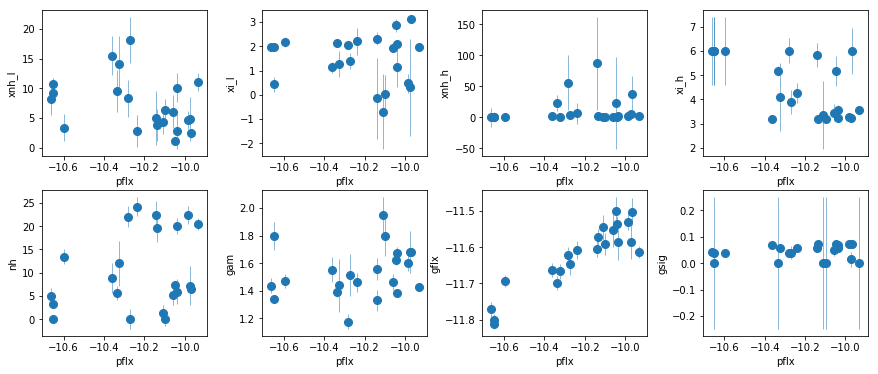
fit_2e: Test adding a low $\xi$ to fit_2d
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_2e = fit_xspec_model('fit_2e', spec_ids, base_dir)
# plot the result #
par_names = ['xnh_l', 'xi_l', 'xnh_h', 'xi_h', 'nh', 'pflx', 'gam', 'gflx', 'gsig']
fit = fit_2e
fig = plt.figure(figsize=(12,5))
idx = [0,1,2,3,4,6,7,8]; iref = 5
for i,ix in enumerate(idx):
ax = plt.subplot(2,len(idx)//2,i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
# write the residuals #
extra_cmd ='del 4; del 5; del 6; freez [az_free]; thaw 12 16; new 20=0.12*16;fit'
write_resid(base_dir, spec_ids, '2e', extra_cmd, -1, avg_bin=True, outdir='results/explore')
fit_2f: Test adding a broad gaussian to fit_2d
os.chdir('%s/%s'%(base_dir, spec_dir))
for i in spec_ids:
os.system("sed -i 's/\*cflux\*powerlaw/\(cflux\*powerlaw\)/g' fits/fit_2d__%d.xcm"%i)
fit_2f = fit_xspec_model('fit_2f', spec_ids, base_dir)
# plot the result #
par_names = ['xnh_h', 'xi_h', 'nh', 'bge', 'bgs', 'bgn', 'pflx', 'gam', 'gflx', 'gsig']
fit = fit_2f
fig = plt.figure(figsize=(10,6))
idx = [0,1,2,3,4,5,7,8,9]; iref = 6
for i,ix in enumerate(idx):
ax = plt.subplot(3,len(idx)//3,i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
# write the residuals #
extra_cmd ='del 4; del 5; del 6; freez [az_free]; thaw 12 16; new 20=0.12*16;fit'
write_resid(base_dir, spec_ids, '2f', extra_cmd, -1, avg_bin=True, outdir='results/explore')
Both options, a low-xi WA (fit_2e) and a broad gaussian (fit_2f) seem to do a good job (the broad line model seems to do slightly better, but it give flat gamma <1.3 for the new datasets). But before deciding, we need to extend the fitting band to lower energies.
fit_2g: Extend fit_2d (neutral+high-$\xi$) to 2.5 keV
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_2g = fit_xspec_model('fit_2g', spec_ids, base_dir)
# plot the result #
par_names = ['xnh', 'xi', 'nh', 'pflx', 'gam', 'gflx', 'gsig']
fit = fit_2g
fig = plt.figure(figsize=(12,3))
idx = [0,1,2,4,5,6]; iref = 3
for i,ix in enumerate(idx):
ax = plt.subplot(1,len(idx),i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
# write the residuals #
extra_cmd ='del 3; del 4; del 5; freez [az_free]; thaw 8 12; new 16=0.12*12;fit'
write_resid(base_dir, spec_ids, '2g', extra_cmd, -1, avg_bin=True, outdir='results/explore')
This model is more of a descriptive one. Gamma<1 for many spectra (mostly the new ones).
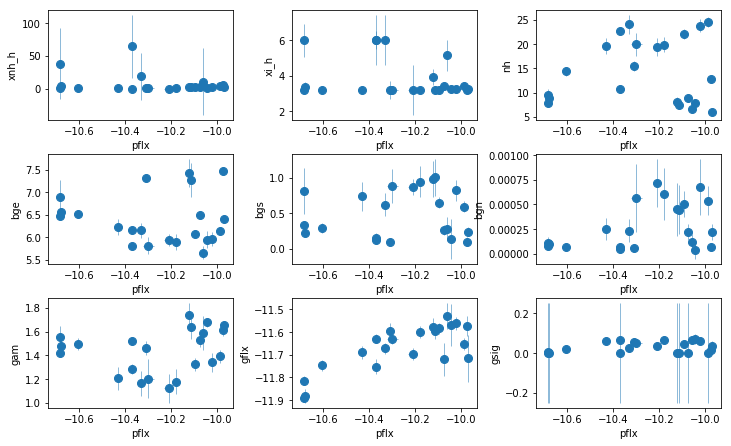
fit_2h: Add an extra PL (PC or scattering)
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_2h = fit_xspec_model('fit_2h', spec_ids, base_dir)
# plot the result #
par_names = ['xnh', 'xi', 'nh', 'pflx', 'gam', 'p2flx', 'gflx', 'gsig']
fit = fit_2h
fig = plt.figure(figsize=(12,6))
idx = [0,1,2,4,5,6,7]; iref = 3
for i,ix in enumerate(idx):
ax = plt.subplot(2,len(idx)//2+1,i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
# write the residuals #
extra_cmd ='del 4; del 5; del 6; del 7; freez [az_free]; thaw 9 11 15; new 19=0.12*15;fit'
write_resid(base_dir, spec_ids, '2h', extra_cmd, -1, avg_bin=True, outdir='results/explore')
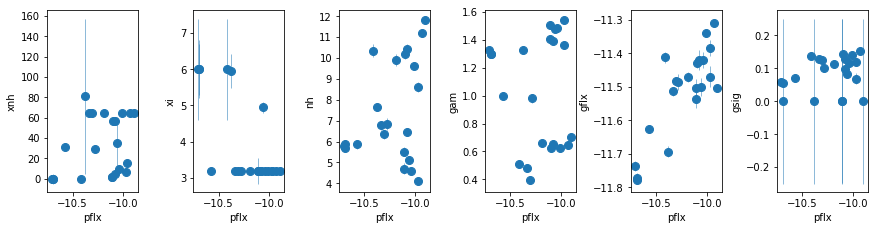
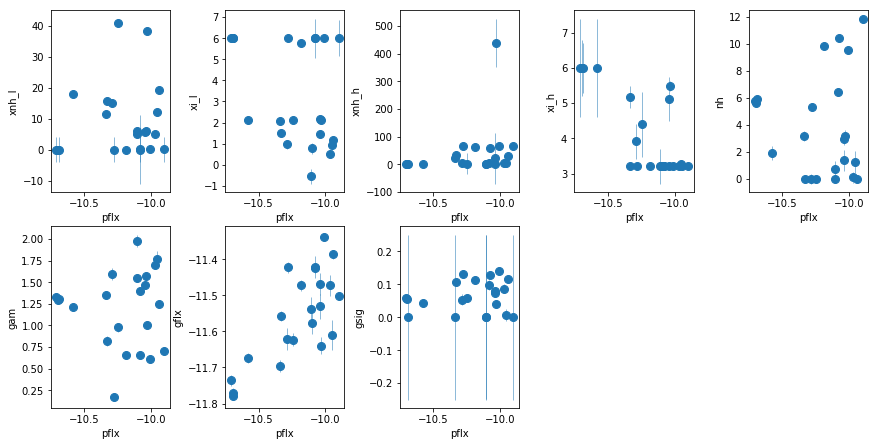
fit_2h1: Use WA instead of the extra PL
Show it doesn’t work
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_2h1 = fit_xspec_model('fit_2h1', spec_ids, base_dir)
# plot the result #
par_names = ['xnh_l', 'xi_l', 'xnh_h', 'xi_h', 'nh', 'pflx', 'gam', 'gflx', 'gsig']
fit = fit_2h1
fig = plt.figure(figsize=(12,6))
idx = [0,1,2,3,4,6,7,8]; iref = 5
for i,ix in enumerate(idx):
ax = plt.subplot(2,len(idx)//2+1,i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
# write the residuals #
extra_cmd ='del 5; del 6; del 7; freez [az_free]; thaw 13 17; new 21=0.12*17;fit'
write_resid(base_dir, spec_ids, '2h1', extra_cmd, -1, avg_bin=True, outdir='results/explore')
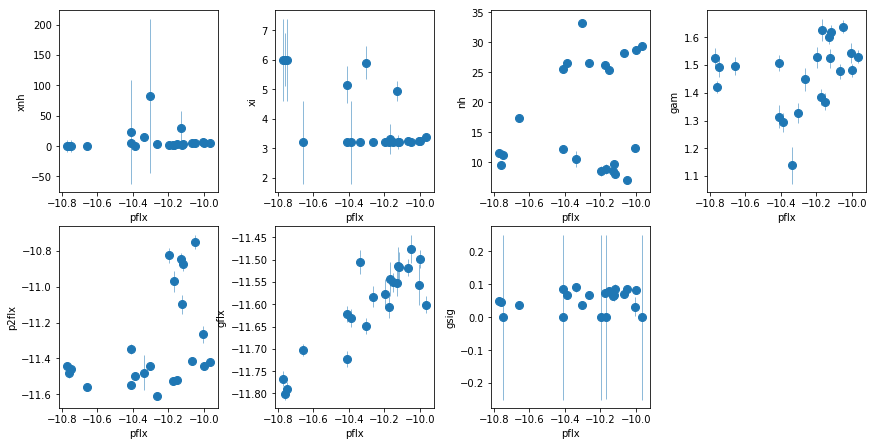
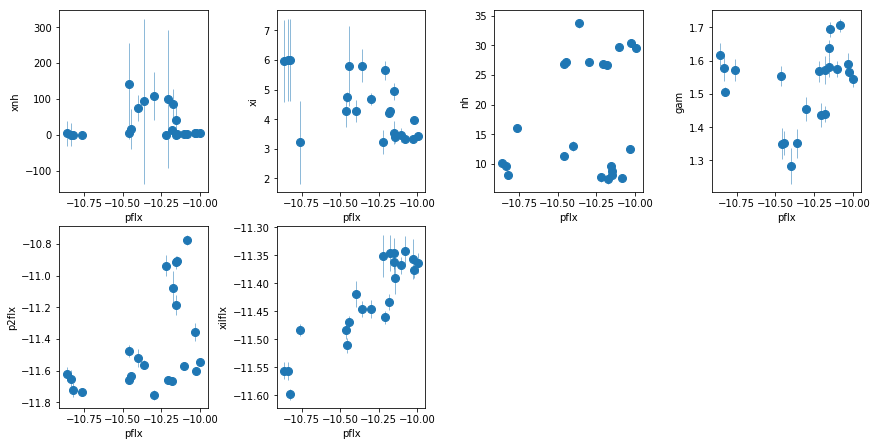
fit_2i: Use xillver for lines, keep the extra PL
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_2i = fit_xspec_model('fit_2i', spec_ids, base_dir)
# plot the result #
par_names = ['xnh', 'xi', 'nh', 'pflx', 'gam', 'p2flx', 'xilflx']
fit = fit_2i
fig = plt.figure(figsize=(12,6))
idx = [0,1,2,4,5,6]; iref = 3
for i,ix in enumerate(idx):
ax = plt.subplot(2,len(idx)//2+1,i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
# write the residuals #
extra_cmd ='del 4; del 5; del 6; freez [az_free]; thaw 9 11 19;fit'
write_resid(base_dir, spec_ids, '2i', extra_cmd, -1, avg_bin=True, outdir='results/explore')
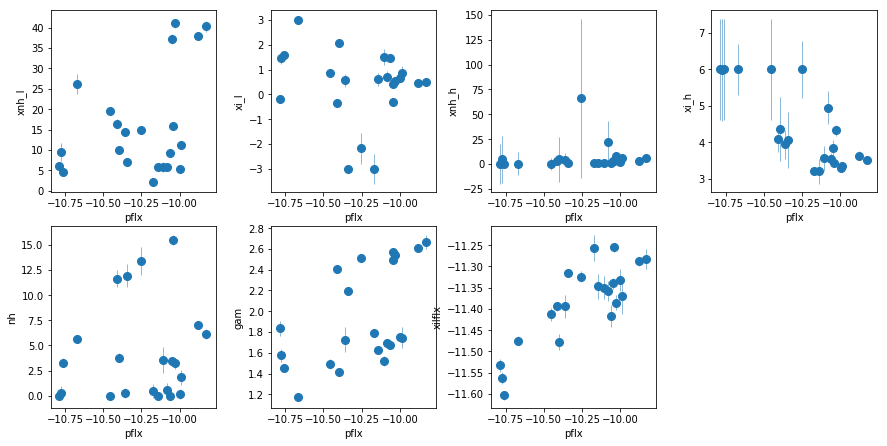
fit_2i1: Use WA instead of extra PL, with xillver
i.e. similar to fit_2h1, but using xillver instead of gaussian emission lines
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_2i1 = fit_xspec_model('fit_2i1', spec_ids, base_dir)
# plot the result #
par_names = ['xnh_l', 'xi_l', 'xnh_h', 'xi_h', 'nh', 'pflx', 'gam', 'xilflx']
fit = fit_2i1
fig = plt.figure(figsize=(12,6))
idx = [0,1,2,3,4,6,7]; iref = 5
for i,ix in enumerate(idx):
ax = plt.subplot(2,len(idx)//2+1,i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
# write the residuals #
extra_cmd ='del 5; del 6; freez [az_free]; thaw 13 21;fit'
write_resid(base_dir, spec_ids, '2i1', extra_cmd, -1, avg_bin=True, outdir='results/explore')
The extra powerlaw is clearly needed (
fit_2hvsfit_2g). A warm absorber does not do the job (fit_2h1). Using xillver instead of narrow lines (fit_2i) first, accouns from some of the broadening of the narrow line, and second, when using a WA instead of the extra powerlaw (fit_2i1), the model is not as bad asfit_2h1because unabsorbed xillver acts like the extra powerlaw. The fits are however leave features around 7-8 keV due to the edge not being modeled correctly
fit_2j: Remove extra PL from fit_2i, and make xillver ionized
To get more soft flux, instead of the WA in fit_2i1
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_2j = fit_xspec_model('fit_2j', spec_ids, base_dir)
# plot the result #
par_names = ['xnh', 'xi', 'nh', 'pflx', 'gam', 'xilflx']
fit = fit_2j
fig = plt.figure(figsize=(12,3))
idx = [0,1,2,4,5]; iref = 3
for i,ix in enumerate(idx):
ax = plt.subplot(1,len(idx),i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
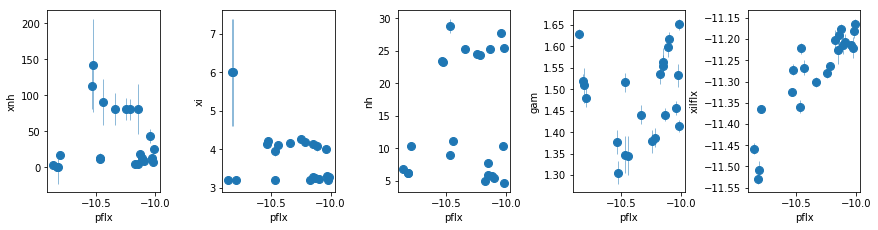
# write the residuals #
extra_cmd ='del 4; del 5; freez [az_free]; thaw 9 17;fit'
write_resid(base_dir, spec_ids, '2j', extra_cmd, -1, avg_bin=True, outdir='results/explore')
Summary so far:
Three possible solutions to the excess:
- Extra PL: using lines
2h, using xillver2i - ionized xillver:
2j(but gamma is small in new data) - Full covering WA: rulled out by
2h1, 2i1
Iron overabundance?
Some early work suggested abundance of x2 solar. It is however not clear how to measure this. Comparing the edge to the line flux is flawed, because the two are not produced by the same material as suggested by their long term variability, where we showed in the narrow line work that they vary totally independently.
Broadband Characterization of the Spectra
Here, we fit descriptive models to all the spectra and track their variability. The model includes:
- absorbed powerlaw and and a narrow line at 6.4 keV for the hard band.
- A soft powerlaw and two lines at 0.56 and 0.9 keV for the soft band.
- A broad gaussian at ~3 keV for the extra complexity.
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_3 = fit_xspec_model('fit_3', spec_ids, base_dir)
# plot the result #
par_names = ['nh', 'hflx', 'hgam', 'g6flx', 'g6s', 'sflx', 'sgam',
'g5flx', 'g5e', 'g9flx', 'g9e', 'gmflx', 'gme', 'gms']
fit = fit_3
fig = plt.figure(figsize=(12,8))
idx = [0,2,3,4,5,6,7,8,9,10,11,12,13]; iref = 1
for i,ix in enumerate(idx):
ax = plt.subplot(3,len(idx)//3+1,i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
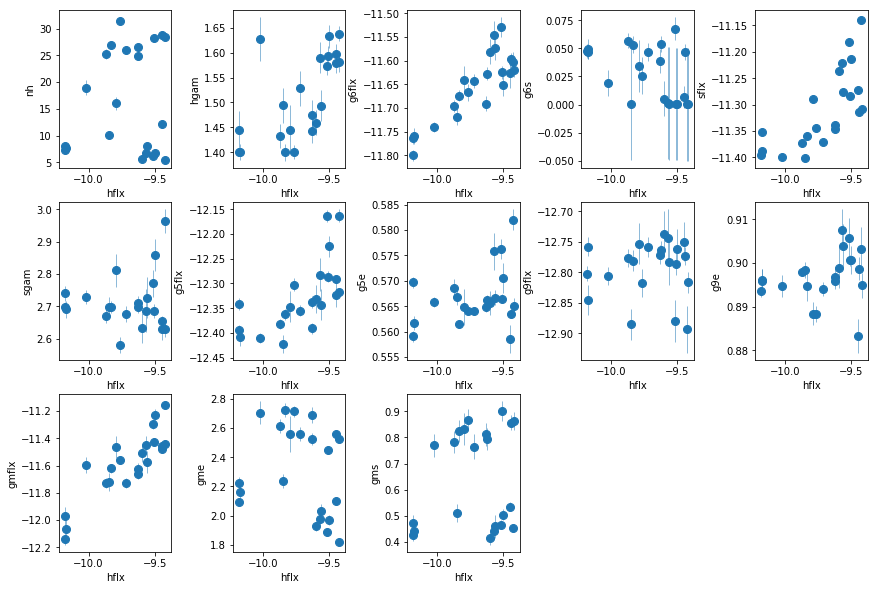
fit_3a: Use PC in the descriptive model instead of a gaussian
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_3a = fit_xspec_model('fit_3a', spec_ids, base_dir)
# plot the result #
par_names = ['nh_f', 'nh', 'cf', 'hflx', 'hgam', 'g6flx', 'g6s', 'sflx', 'sgam',
'g5flx', 'g5e', 'g9flx', 'g9e']
fit = fit_3a
fig = plt.figure(figsize=(12,8))
idx = [0,1,2,4,5,6,7,8,9,10,11,12]; iref = 3
for i,ix in enumerate(idx):
ax = plt.subplot(3,len(idx)//3+1,i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
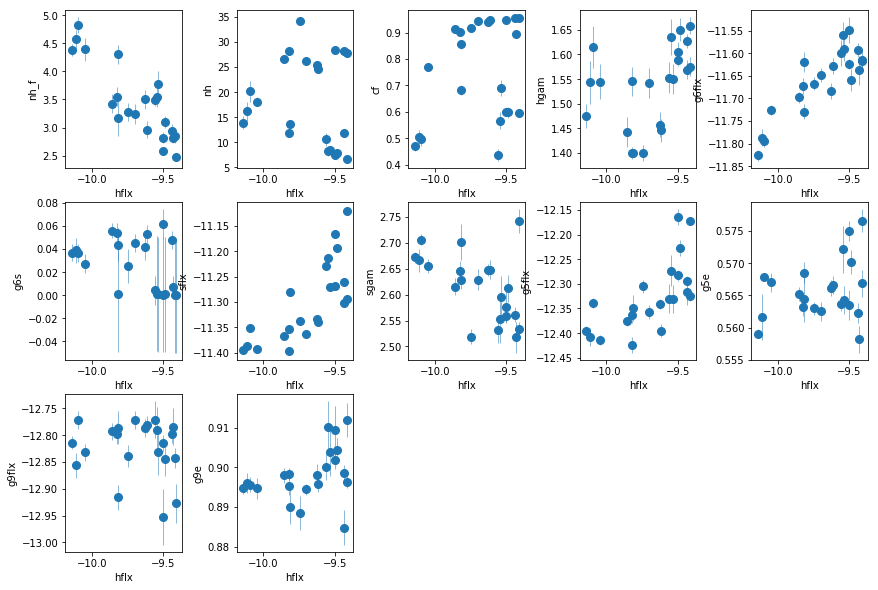
nh_fis anti correlated with hard flux. THIS IS IT. Evidence for WA.
fit_3a1: Similar to fit_3a, but constrain Cf>0.9
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_3a1 = fit_xspec_model('fit_3a1', spec_ids, base_dir)
# plot the result #
par_names = ['nh_f', 'nh', 'cf', 'hflx', 'hgam', 'g6flx', 'g6s', 'sflx', 'sgam',
'g5flx', 'g5e', 'g9flx', 'g9e']
fit = fit_3a1
fig = plt.figure(figsize=(12,8))
idx = [0,1,2,4,5,6,7,8,9,10,11,12]; iref = 3
for i,ix in enumerate(idx):
ax = plt.subplot(3,len(idx)//3+1,i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
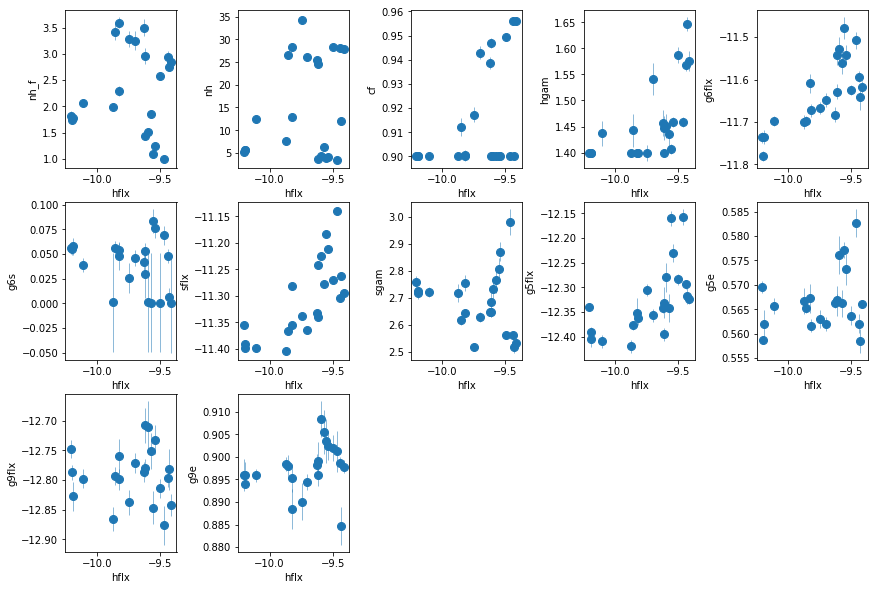
fit_3b: Make the full absorber ionized
Because of the degenrecy between nh and cf, limit the nh of the main absorber to a maximum of 40, as observed when the full absorber is neutral
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_3b = fit_xspec_model('fit_3b', spec_ids, base_dir)
# plot the result #
par_names = ['xnh', 'xi', 'nh', 'cf', 'hflx', 'hgam', 'g6flx', 'g6s', 'sflx', 'sgam',
'g5flx', 'g5e', 'g9flx', 'g9e']
fit = fit_3b
fig = plt.figure(figsize=(12,8))
idx = [0,1,2,3,5,6,7,8,9,10,11,12,13]; iref = 4
for i,ix in enumerate(idx):
ax = plt.subplot(3,len(idx)//3+1,i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
nh is somwhat less correlated with flux now. xi of the added WA appears to be correlated with flux??
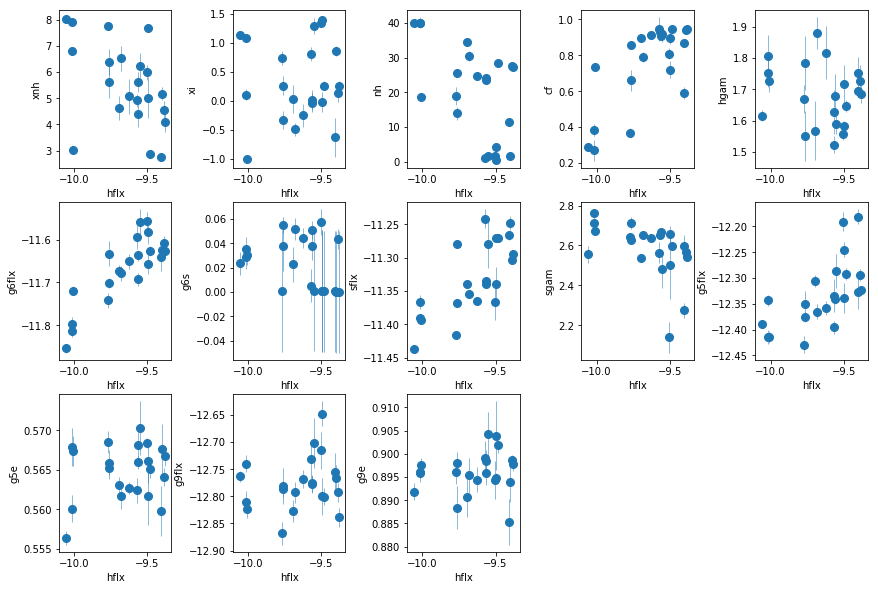
fit_3c: Similar to fit_3b, fixing full absorber
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_3c = fit_xspec_model('fit_3c', spec_ids, base_dir)
# plot the result #
par_names = ['xnh', 'xi', 'hflx', 'hgam', 'g6flx', 'g6s', 'sflx', 'sgam',
'g5flx', 'g5e', 'g9flx', 'g9e']
fit = fit_3c
fig = plt.figure(figsize=(12,8))
idx = [0,1,3,4,5,6,7,8,9,10,11]; iref = 2
for i,ix in enumerate(idx):
ax = plt.subplot(3,len(idx)//3+1,i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
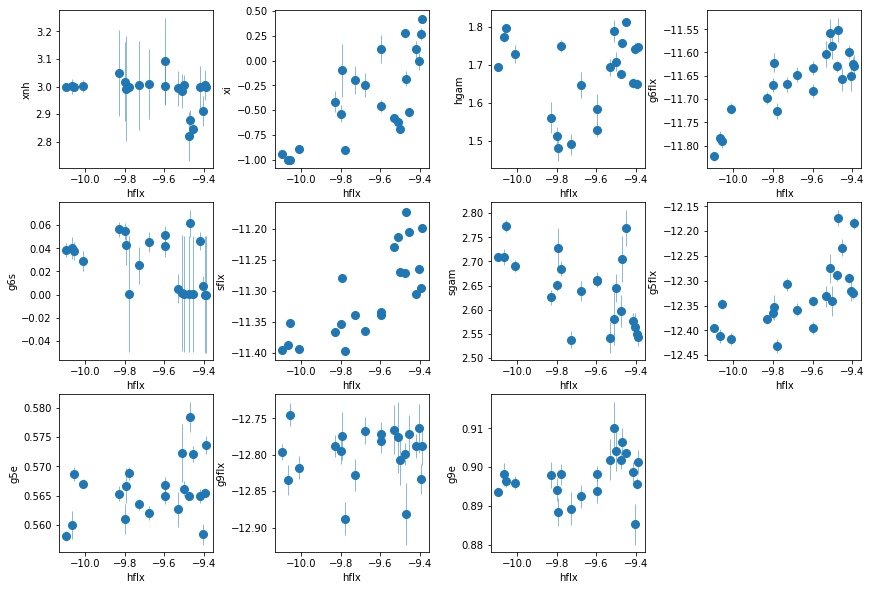
The trends are clearer now. The Nh of WA is constant, and its ionization changes with flux
fit_3d: Leaked xillver; similar to 2i1 but 0.3-10 keV
os.chdir('%s/%s'%(base_dir, spec_dir))
fit_3d = fit_xspec_model('fit_3c', spec_ids, base_dir)
# plot the result #
par_names = ['xnh', 'xi', 'nh', 'hflx', 'hgam', 'xilflx', 'xilxi',
'g5flx', 'g5e', 'g9flx', 'g9e']
fit = fit_3d
fig = plt.figure(figsize=(12,8))
idx = [0,1,2,4,5,6,7,8,9,10]; iref = 3
for i,ix in enumerate(idx):
ax = plt.subplot(3,len(idx)//3+1,i+1)
plt.errorbar(fit[:,iref,0], fit[:,ix,0], fit[:,ix,1], xerr=fit[:,iref,1],
fmt='o', ms=8, lw=0.5)
ax.set_xlabel(par_names[iref]); ax.set_ylabel(par_names[ix])
plt.tight_layout(pad=0)
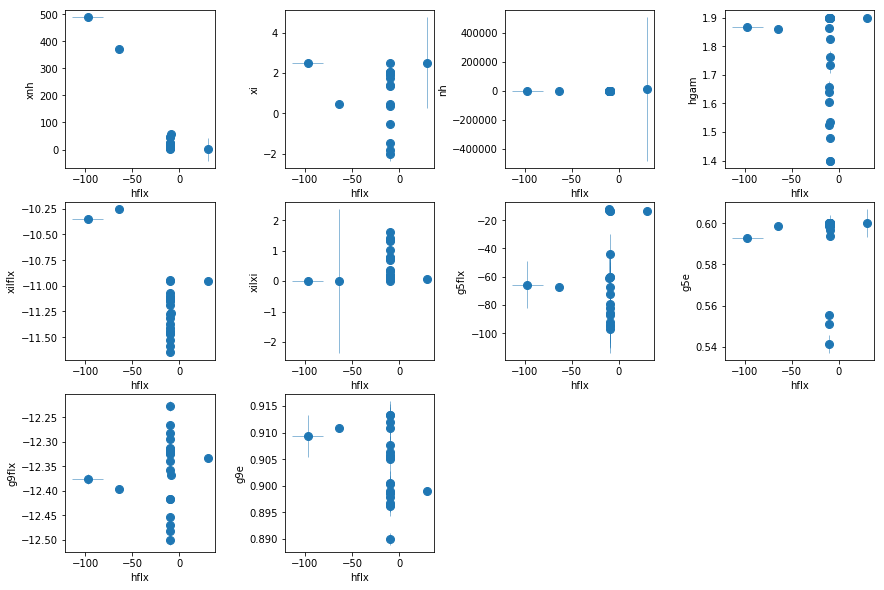
The fit is not good in this case. The soft spectra need a steeper model then what is needed in the hard spectrum
Summary plot with nh vs flux
# plot nh vs hflux with numbers
fig = plt.figure(figsize=(6,6))
plt.plot(fit_3[:,1,0], fit_3[:,0,0], 'o', ms=30, markerfacecolor='none')
#plt.text?
for i,s in enumerate(spec_ids):
plt.text(fit_3[i,1,0], fit_3[i,0,0], '%d'%s, horizontalalignment='center',
verticalalignment='center')
#(fit_2[:,1,0], fit_2[:,0,0], ['%d'%i for i in spec_ids])
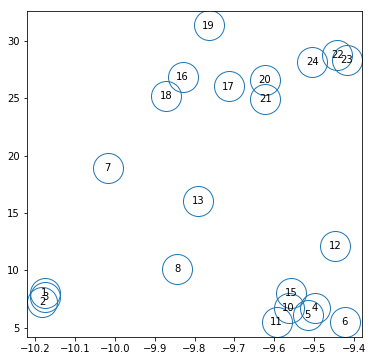
The respresentitave spectra based on flux, nh and exposure we use are: 2, 6, 7, 16, 23
Soft variability: Evidence for Warm absorber
Pick two sets of observation pairs where nh is roughly constant but the soft band changes, and calculate difference spectra. We need the pairs to be close in time so the gain doesn’t change a lot, i.e. it is safe to assume the same rmf file when doing the difference in xspec
# sort the observations by soft flux; and print the nh along with that sorting
isort_soft = np.argsort(fit_3[:,5,0])[::-1]
print('{:5} {:5} {:5}'.format('id', 'sflx', 'nh'))
for i in isort_soft:
print('{:5} {:5.4} {:5.3}'.format(spec_ids[i], fit_3[i,6,0], fit_3[i,0,0]))
id sflx nh
6 2.963 5.51
5 2.771 6.12
4 2.86 6.75
10 2.686 6.69
11 2.632 5.53
12 2.629 12.1
15 2.727 8.02
24 2.686 28.2
13 2.812 16.0
23 2.631 28.4
22 2.654 28.8
20 2.698 26.6
19 2.582 31.3
21 2.71 25.0
1 2.741 8.02
16 2.697 26.8
17 2.676 26.0
18 2.671 25.2
3 2.691 7.68
2 2.698 7.26
7 2.73 18.9
8 2.699 10.1
We do the following pairs: 6-4, 23-20
# soft_variability_diff
os.chdir('%s/%s'%(base_dir, spec_dir))
ispec = [[6, 4], [23, 20]]
diff_soft = []
for ih,il in ispec:
tcl = 'source %s/fit.tcl\n'%base_dir
tcl += 'soft_variability_diff %d %d\nexit\n'%(ih, il)
xcm = 'tmp_lh.xcm'
with open(xcm, 'w') as fp: fp.write(tcl)
cmd = 'xspec - %s > tmp_lh.log 2>&1'%(xcm)
p = subp.call(['/bin/bash', '-i', '-c', cmd])
spec = [np.loadtxt('tmp_%s.dat'%x, skiprows=3) for x in ['l', 'h', 'd']]
diff_soft.append(spec)
_ = os.system('rm tmp_[l,h,d].dat tmp_lh.*')
# write the results to a veusz file for plotting #
text = ''
for i,res in enumerate(diff_soft):
for il,l in enumerate(['l', 'h']):
text += '\ndescriptor en_{0}_i{1},+- d_{0}_i{1},+-\n'.format(l, i+1)
text += '\n'.join(['{} {} {} {}'.format(*x) for x in res[il]])
text += ('\ndescriptor en_diff_i{0},+- d_diff_i{0},+- mtot_diff_i{0} '
'm1_diff_i{0} m2_diff_i{0}\n').format(i+1)
text += '\n'.join(['{0} {1} {2} {3} {4} {5} {6}'.format(
res[2][i,0], res[2][i,1], res[2][i,2], res[2][i,3],
res[2][i,4], res[2][i,5], res[2][i,6])
for i in range(len(res[2]))])
with open('results/explore/soft_variability_diff.plot', 'w') as fp: fp.write(text)
Hard Spectrum: The need for Partial Covering
Use observation 24 to illustrate the need for partial covering
os.chdir('%s/%s'%(base_dir, spec_dir))
tcl = 'source %s/fit.tcl\n'%base_dir
tcl += 'fit_hard_pc spec_24.grp\nexit\n'
with open('tmp_h.xcm', 'w') as fp: fp.write(tcl)
cmd = 'xspec - tmp_h.xcm > tmp_h.log 2>&1'
p = subp.call(['/bin/bash', '-i', '-c', cmd])
# 6(cases: full,pc,fe,wa,ga,po), nen, 5 (en,+-,d,+-,m,mc1,mc2...)
hard_spec = [np.loadtxt('tmp_%d.dat'%x, skiprows=3) for x in range(1,7)]
_ = os.system('rm tmp_?.dat')
text = ''
for ih,hs in enumerate(hard_spec):
text += '\ndescriptor en_s{0},+- d_s{0},+- mtot_s{0} res_s{0},+-\n'.format(ih+1)
text += '\n'.join(['{} {} {} {} {} {} {}'.format(
x[0], x[1], x[2], x[3], x[4], (x[2]-x[4])/x[3], 1.0) for x in hs])
text += '\ndescriptor %s\n'%(' '.join(['mc%d_s%d'%(ic+1, ih+1) for ic in range(hs.shape[1]-5)]))
text += '\n'.join([' '.join(['%g'%xx for xx in x[5:]]) for x in hs])
with open('results/explore/hard_pc.plot', 'w') as fp: fp.write(text)